| Full name: proline rich 26 | Alias Symbol: FLJ38681 | ||
| Type: protein-coding gene | Cytoband: 10p15.3 | ||
| Entrez ID: 414235 | HGNC ID: HGNC:30724 | Ensembl Gene: ENSG00000180525 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of PRR26:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRR26 | 414235 | 1559000_at | -0.4293 | 0.4052 | |
| GSE26886 | PRR26 | 414235 | 1559000_at | 0.2570 | 0.0251 | |
| GSE45670 | PRR26 | 414235 | 1559000_at | -0.3285 | 0.0001 | |
| GSE53622 | PRR26 | 414235 | 11043 | -2.6142 | 0.0000 | |
| GSE53624 | PRR26 | 414235 | 11043 | -2.3402 | 0.0000 | |
| GSE63941 | PRR26 | 414235 | 1559000_at | -0.0476 | 0.7686 | |
| GSE77861 | PRR26 | 414235 | 1559000_at | 0.0090 | 0.9620 | |
| SRP133303 | PRR26 | 414235 | RNAseq | -0.1924 | 0.5034 | |
| SRP159526 | PRR26 | 414235 | RNAseq | -1.7256 | 0.0525 | |
| SRP193095 | PRR26 | 414235 | RNAseq | 0.2764 | 0.3077 | |
| SRP219564 | PRR26 | 414235 | RNAseq | -0.8925 | 0.2944 |
Upregulated datasets: 0; Downregulated datasets: 2.
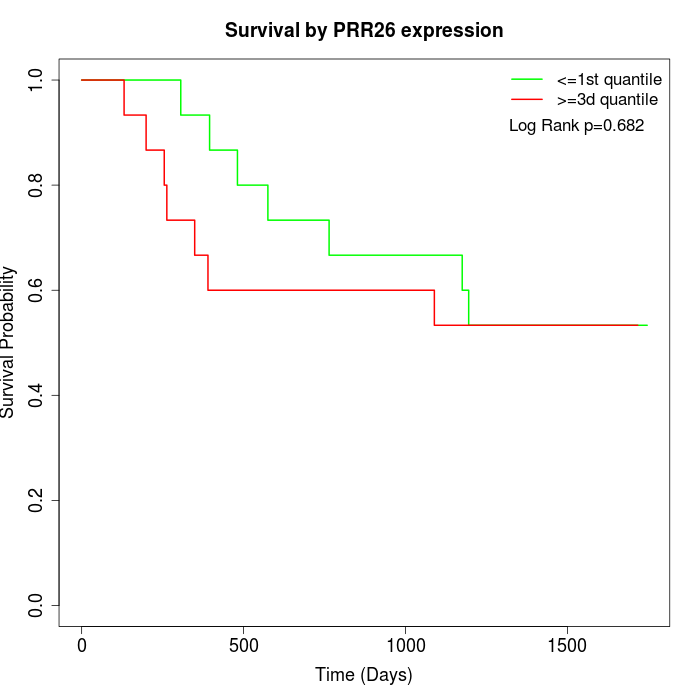
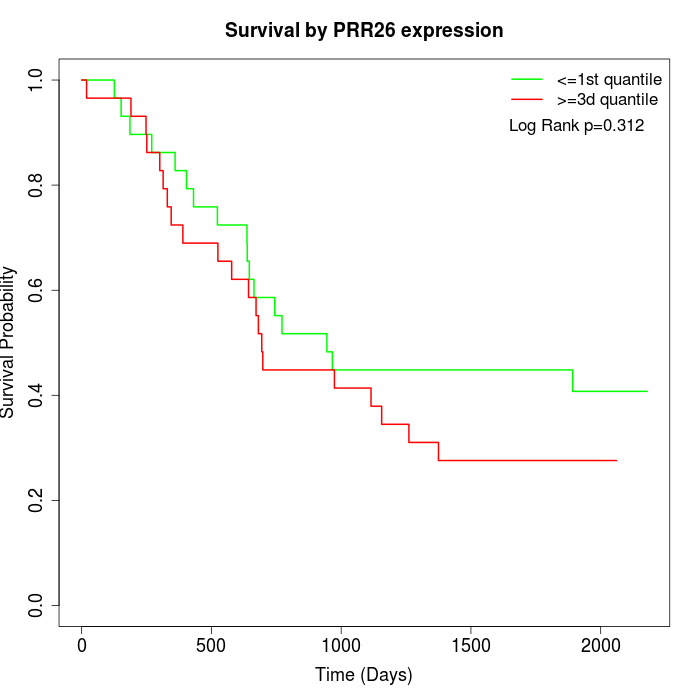
Survival by PRR26 expression:
Note: Click image to view full size file.
Copy number change of PRR26:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRR26 | 414235 | 5 | 10 | 15 | |
| GSE20123 | PRR26 | 414235 | 5 | 10 | 15 | |
| GSE43470 | PRR26 | 414235 | 3 | 8 | 32 | |
| GSE46452 | PRR26 | 414235 | 2 | 14 | 43 | |
| GSE47630 | PRR26 | 414235 | 5 | 15 | 20 | |
| GSE54993 | PRR26 | 414235 | 11 | 0 | 59 | |
| GSE54994 | PRR26 | 414235 | 4 | 10 | 39 | |
| GSE60625 | PRR26 | 414235 | 0 | 0 | 11 | |
| GSE74703 | PRR26 | 414235 | 2 | 5 | 29 | |
| GSE74704 | PRR26 | 414235 | 1 | 9 | 10 | |
| TCGA | PRR26 | 414235 | 18 | 26 | 52 |
Total number of gains: 56; Total number of losses: 107; Total Number of normals: 325.
Somatic mutations of PRR26:
Generating mutation plots.
Highly correlated genes for PRR26:
Showing top 20/491 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRR26 | FRRS1L | 0.844217 | 3 | 0 | 3 |
| PRR26 | MYOM1 | 0.822422 | 4 | 0 | 4 |
| PRR26 | FENDRR | 0.81816 | 4 | 0 | 4 |
| PRR26 | PYGM | 0.815544 | 4 | 0 | 4 |
| PRR26 | ANGPTL1 | 0.810508 | 4 | 0 | 4 |
| PRR26 | ACACB | 0.808705 | 3 | 0 | 3 |
| PRR26 | PPP1R12B | 0.80605 | 4 | 0 | 4 |
| PRR26 | TTLL7 | 0.793228 | 3 | 0 | 3 |
| PRR26 | LMOD1 | 0.791809 | 4 | 0 | 4 |
| PRR26 | MYOCD | 0.789368 | 4 | 0 | 4 |
| PRR26 | ASB5 | 0.78722 | 4 | 0 | 4 |
| PRR26 | DES | 0.782024 | 4 | 0 | 4 |
| PRR26 | BHMT2 | 0.778939 | 4 | 0 | 4 |
| PRR26 | CDH19 | 0.775959 | 4 | 0 | 4 |
| PRR26 | KCNMB1 | 0.775521 | 4 | 0 | 4 |
| PRR26 | CACNB2 | 0.774858 | 5 | 0 | 5 |
| PRR26 | CCDC69 | 0.774621 | 4 | 0 | 3 |
| PRR26 | NEGR1 | 0.77395 | 4 | 0 | 4 |
| PRR26 | PDE5A | 0.769557 | 4 | 0 | 4 |
| PRR26 | CTNNA3 | 0.765678 | 4 | 0 | 4 |
For details and further investigation, click here