| Full name: acetylcholinesterase (Cartwright blood group) | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 43 | HGNC ID: HGNC:108 | Ensembl Gene: ENSG00000087085 | OMIM ID: 100740 |
| Drug and gene relationship at DGIdb | |||
Expression of ACHE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACHE | 43 | 210332_at | -0.0301 | 0.9140 | |
| GSE20347 | ACHE | 43 | 210332_at | -0.0824 | 0.3413 | |
| GSE23400 | ACHE | 43 | 210332_at | -0.2152 | 0.0000 | |
| GSE26886 | ACHE | 43 | 210332_at | 0.2936 | 0.0209 | |
| GSE29001 | ACHE | 43 | 205377_s_at | -0.0495 | 0.7885 | |
| GSE38129 | ACHE | 43 | 210332_at | -0.0837 | 0.2289 | |
| GSE45670 | ACHE | 43 | 210332_at | 0.0817 | 0.4178 | |
| GSE53622 | ACHE | 43 | 26410 | -0.0232 | 0.9203 | |
| GSE53624 | ACHE | 43 | 26410 | -0.7921 | 0.0000 | |
| GSE63941 | ACHE | 43 | 210332_at | 0.1378 | 0.3209 | |
| GSE77861 | ACHE | 43 | 210332_at | -0.0681 | 0.7409 | |
| GSE97050 | ACHE | 43 | A_24_P60845 | 0.1952 | 0.5885 | |
| SRP064894 | ACHE | 43 | RNAseq | -0.6960 | 0.0179 | |
| SRP133303 | ACHE | 43 | RNAseq | -0.3575 | 0.3769 | |
| SRP159526 | ACHE | 43 | RNAseq | -0.2832 | 0.5377 | |
| SRP219564 | ACHE | 43 | RNAseq | -1.0740 | 0.0222 | |
| TCGA | ACHE | 43 | RNAseq | -0.3529 | 0.1708 |
Upregulated datasets: 0; Downregulated datasets: 1.
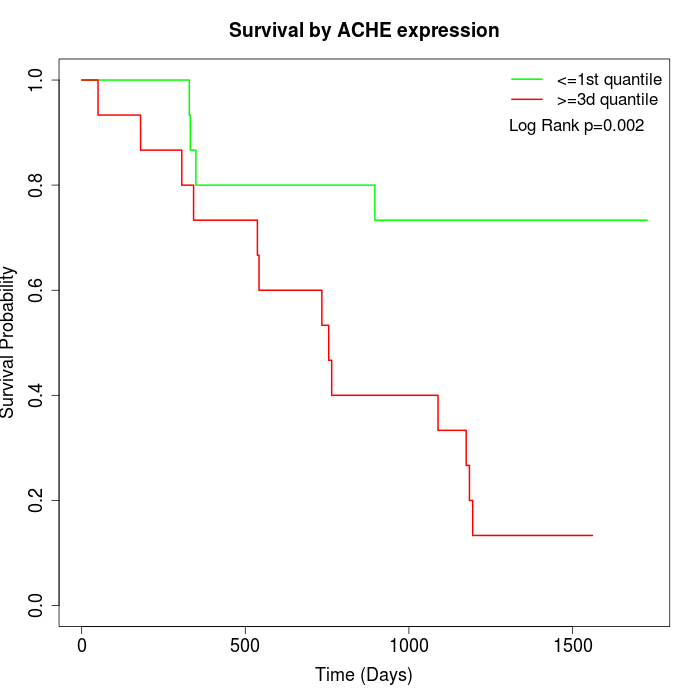
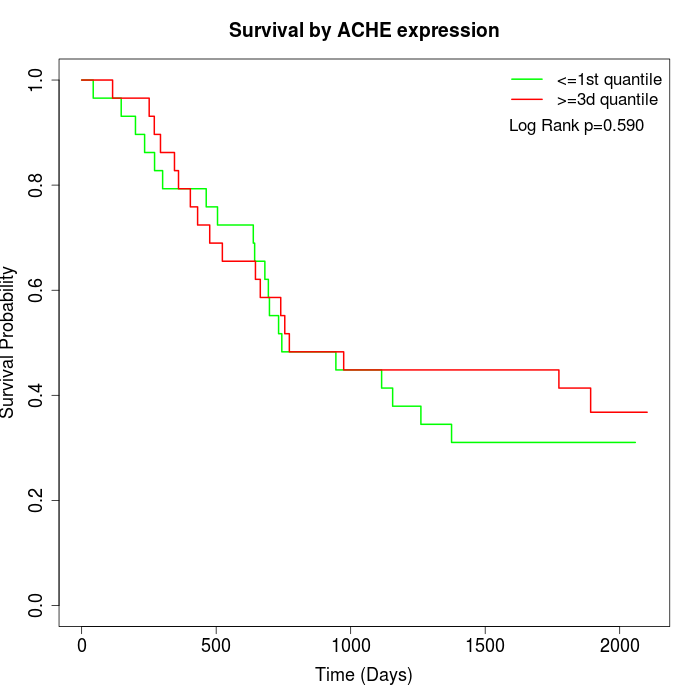
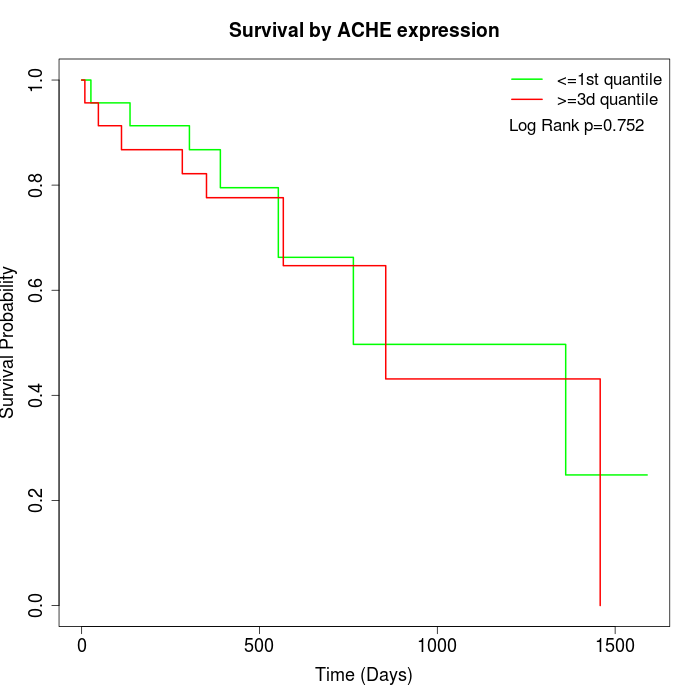
Survival by ACHE expression:
Note: Click image to view full size file.
Copy number change of ACHE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACHE | 43 | 13 | 0 | 17 | |
| GSE20123 | ACHE | 43 | 13 | 0 | 17 | |
| GSE43470 | ACHE | 43 | 7 | 2 | 34 | |
| GSE46452 | ACHE | 43 | 11 | 1 | 47 | |
| GSE47630 | ACHE | 43 | 7 | 3 | 30 | |
| GSE54993 | ACHE | 43 | 1 | 9 | 60 | |
| GSE54994 | ACHE | 43 | 15 | 3 | 35 | |
| GSE60625 | ACHE | 43 | 0 | 0 | 11 | |
| GSE74703 | ACHE | 43 | 7 | 1 | 28 | |
| GSE74704 | ACHE | 43 | 9 | 0 | 11 | |
| TCGA | ACHE | 43 | 53 | 6 | 37 |
Total number of gains: 136; Total number of losses: 25; Total Number of normals: 327.
Somatic mutations of ACHE:
Generating mutation plots.
Highly correlated genes for ACHE:
Showing top 20/611 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACHE | WDR27 | 0.801623 | 3 | 0 | 3 |
| ACHE | APOC2 | 0.785635 | 3 | 0 | 3 |
| ACHE | PPME1 | 0.693375 | 3 | 0 | 3 |
| ACHE | STRADA | 0.690054 | 3 | 0 | 3 |
| ACHE | KLC1 | 0.689691 | 4 | 0 | 4 |
| ACHE | KRTAP4-3 | 0.677337 | 3 | 0 | 3 |
| ACHE | DLG5-AS1 | 0.672536 | 3 | 0 | 3 |
| ACHE | DAB2IP | 0.669403 | 3 | 0 | 3 |
| ACHE | SLC39A3 | 0.668488 | 3 | 0 | 3 |
| ACHE | ZNF676 | 0.668185 | 3 | 0 | 3 |
| ACHE | WDR64 | 0.665289 | 3 | 0 | 3 |
| ACHE | FOXD2-AS1 | 0.664994 | 3 | 0 | 3 |
| ACHE | ROMO1 | 0.661048 | 3 | 0 | 3 |
| ACHE | PLIN4 | 0.658218 | 3 | 0 | 3 |
| ACHE | ADAMTS14 | 0.653305 | 4 | 0 | 3 |
| ACHE | SZT2 | 0.652664 | 4 | 0 | 4 |
| ACHE | UNC5A | 0.651761 | 6 | 0 | 5 |
| ACHE | NACC1 | 0.647587 | 3 | 0 | 3 |
| ACHE | ICAM5 | 0.646388 | 5 | 0 | 5 |
| ACHE | PHLDB1 | 0.645325 | 6 | 0 | 5 |
For details and further investigation, click here