| Full name: ADAM metallopeptidase domain 10 | Alias Symbol: kuz|MADM|HsT18717|CD156C | ||
| Type: protein-coding gene | Cytoband: 15q21.3 | ||
| Entrez ID: 102 | HGNC ID: HGNC:188 | Ensembl Gene: ENSG00000137845 | OMIM ID: 602192 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ADAM10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ADAM10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADAM10 | 102 | 202603_at | 0.3110 | 0.4701 | |
| GSE20347 | ADAM10 | 102 | 202603_at | 0.2509 | 0.0233 | |
| GSE23400 | ADAM10 | 102 | 202603_at | 0.5201 | 0.0000 | |
| GSE26886 | ADAM10 | 102 | 202603_at | 0.6861 | 0.0066 | |
| GSE29001 | ADAM10 | 102 | 202603_at | 0.3401 | 0.1629 | |
| GSE38129 | ADAM10 | 102 | 202603_at | 0.4418 | 0.0000 | |
| GSE45670 | ADAM10 | 102 | 202603_at | 0.3839 | 0.0095 | |
| GSE53622 | ADAM10 | 102 | 55098 | 0.4272 | 0.0000 | |
| GSE53624 | ADAM10 | 102 | 55098 | 0.1588 | 0.1086 | |
| GSE63941 | ADAM10 | 102 | 202603_at | -0.2913 | 0.2604 | |
| GSE77861 | ADAM10 | 102 | 202603_at | 0.4933 | 0.0431 | |
| GSE97050 | ADAM10 | 102 | A_33_P3243857 | 0.5768 | 0.2841 | |
| SRP007169 | ADAM10 | 102 | RNAseq | -0.0164 | 0.9577 | |
| SRP008496 | ADAM10 | 102 | RNAseq | 0.3083 | 0.0661 | |
| SRP064894 | ADAM10 | 102 | RNAseq | 0.3530 | 0.0866 | |
| SRP133303 | ADAM10 | 102 | RNAseq | 0.6137 | 0.0000 | |
| SRP159526 | ADAM10 | 102 | RNAseq | -0.1788 | 0.6070 | |
| SRP193095 | ADAM10 | 102 | RNAseq | -0.0158 | 0.8940 | |
| SRP219564 | ADAM10 | 102 | RNAseq | 0.0683 | 0.8537 | |
| TCGA | ADAM10 | 102 | RNAseq | 0.1342 | 0.0097 |
Upregulated datasets: 0; Downregulated datasets: 0.
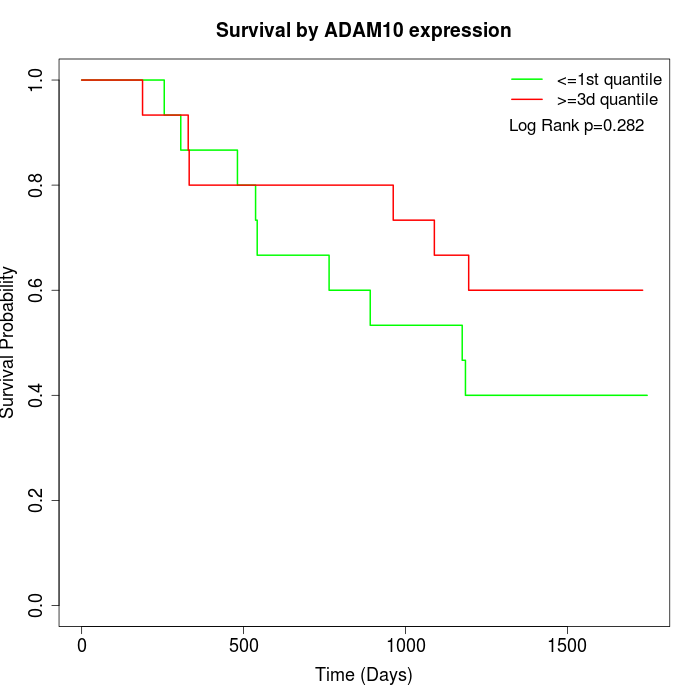
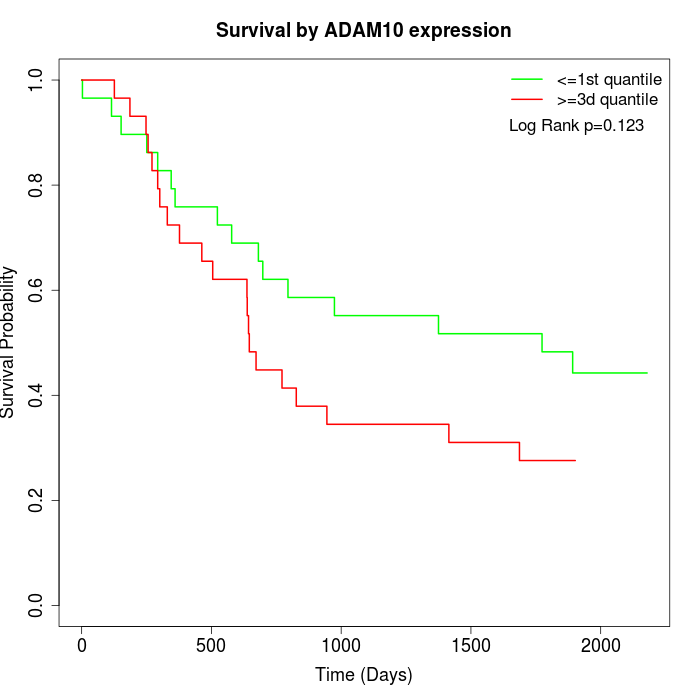
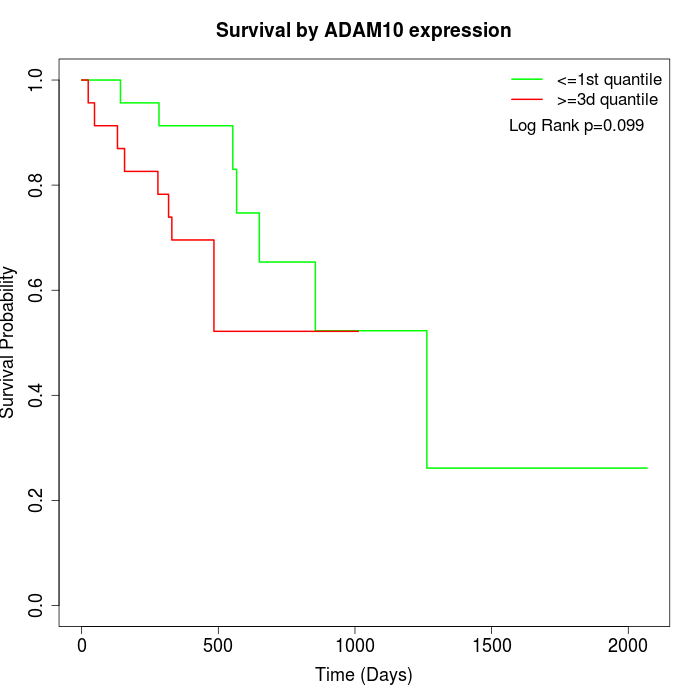
Survival by ADAM10 expression:
Note: Click image to view full size file.
Copy number change of ADAM10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADAM10 | 102 | 6 | 4 | 20 | |
| GSE20123 | ADAM10 | 102 | 6 | 3 | 21 | |
| GSE43470 | ADAM10 | 102 | 4 | 4 | 35 | |
| GSE46452 | ADAM10 | 102 | 3 | 7 | 49 | |
| GSE47630 | ADAM10 | 102 | 8 | 10 | 22 | |
| GSE54993 | ADAM10 | 102 | 5 | 6 | 59 | |
| GSE54994 | ADAM10 | 102 | 6 | 8 | 39 | |
| GSE60625 | ADAM10 | 102 | 4 | 0 | 7 | |
| GSE74703 | ADAM10 | 102 | 4 | 3 | 29 | |
| GSE74704 | ADAM10 | 102 | 2 | 3 | 15 | |
| TCGA | ADAM10 | 102 | 10 | 17 | 69 |
Total number of gains: 58; Total number of losses: 65; Total Number of normals: 365.
Somatic mutations of ADAM10:
Generating mutation plots.
Highly correlated genes for ADAM10:
Showing top 20/1056 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADAM10 | ZNF121 | 0.855151 | 3 | 0 | 3 |
| ADAM10 | INTS10 | 0.79347 | 3 | 0 | 3 |
| ADAM10 | ANKRD50 | 0.76278 | 4 | 0 | 4 |
| ADAM10 | PPP6R3 | 0.745006 | 4 | 0 | 4 |
| ADAM10 | DHX40 | 0.743808 | 3 | 0 | 3 |
| ADAM10 | SPRED2 | 0.73247 | 3 | 0 | 3 |
| ADAM10 | CDCA2 | 0.724293 | 4 | 0 | 4 |
| ADAM10 | SPRED1 | 0.722098 | 6 | 0 | 5 |
| ADAM10 | CARD10 | 0.716738 | 4 | 0 | 3 |
| ADAM10 | PSMG3 | 0.716508 | 5 | 0 | 5 |
| ADAM10 | NOP58 | 0.715825 | 4 | 0 | 4 |
| ADAM10 | ZNF627 | 0.713293 | 3 | 0 | 3 |
| ADAM10 | ALDH1A3 | 0.710302 | 3 | 0 | 3 |
| ADAM10 | WDR81 | 0.707507 | 3 | 0 | 3 |
| ADAM10 | LASP1 | 0.700864 | 4 | 0 | 4 |
| ADAM10 | UBXN2A | 0.699385 | 3 | 0 | 3 |
| ADAM10 | ANKS6 | 0.697641 | 3 | 0 | 3 |
| ADAM10 | SMU1 | 0.694909 | 4 | 0 | 3 |
| ADAM10 | SLC35A5 | 0.691714 | 4 | 0 | 3 |
| ADAM10 | PGM2L1 | 0.6859 | 3 | 0 | 3 |
For details and further investigation, click here