| Full name: alcohol dehydrogenase 1A (class I), alpha polypeptide | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q23 | ||
| Entrez ID: 124 | HGNC ID: HGNC:249 | Ensembl Gene: ENSG00000187758 | OMIM ID: 103700 |
| Drug and gene relationship at DGIdb | |||
Expression of ADH1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADH1A | 124 | 207820_at | -0.0807 | 0.7753 | |
| GSE20347 | ADH1A | 124 | 207820_at | -0.0189 | 0.8134 | |
| GSE23400 | ADH1A | 124 | 207820_at | -0.0542 | 0.0641 | |
| GSE26886 | ADH1A | 124 | 207820_at | -0.1623 | 0.2127 | |
| GSE29001 | ADH1A | 124 | 207820_at | -0.0923 | 0.4697 | |
| GSE38129 | ADH1A | 124 | 207820_at | -0.2467 | 0.0146 | |
| GSE45670 | ADH1A | 124 | 207820_at | -0.2668 | 0.0194 | |
| GSE63941 | ADH1A | 124 | 207820_at | -0.0450 | 0.8527 | |
| GSE77861 | ADH1A | 124 | 207820_at | -0.1030 | 0.3889 | |
| GSE97050 | ADH1A | 124 | A_24_P291658 | -1.2414 | 0.1291 | |
| SRP133303 | ADH1A | 124 | RNAseq | -0.9368 | 0.0891 | |
| TCGA | ADH1A | 124 | RNAseq | 0.9248 | 0.3522 |
Upregulated datasets: 0; Downregulated datasets: 0.
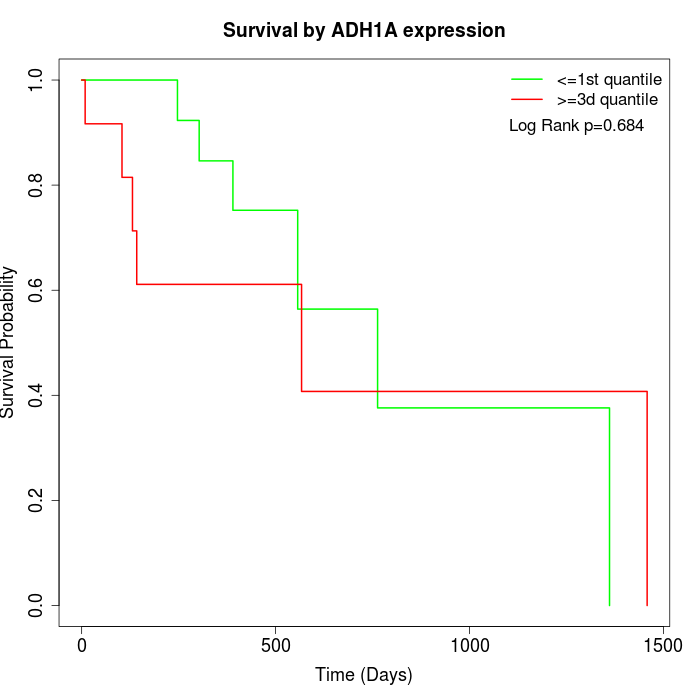
Survival by ADH1A expression:
Note: Click image to view full size file.
Copy number change of ADH1A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADH1A | 124 | 0 | 12 | 18 | |
| GSE20123 | ADH1A | 124 | 0 | 12 | 18 | |
| GSE43470 | ADH1A | 124 | 0 | 14 | 29 | |
| GSE46452 | ADH1A | 124 | 1 | 36 | 22 | |
| GSE47630 | ADH1A | 124 | 0 | 20 | 20 | |
| GSE54993 | ADH1A | 124 | 9 | 0 | 61 | |
| GSE54994 | ADH1A | 124 | 1 | 11 | 41 | |
| GSE60625 | ADH1A | 124 | 0 | 3 | 8 | |
| GSE74703 | ADH1A | 124 | 0 | 12 | 24 | |
| GSE74704 | ADH1A | 124 | 0 | 6 | 14 | |
| TCGA | ADH1A | 124 | 8 | 41 | 47 |
Total number of gains: 19; Total number of losses: 167; Total Number of normals: 302.
Somatic mutations of ADH1A:
Generating mutation plots.
Highly correlated genes for ADH1A:
Showing top 20/275 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADH1A | TRIM3 | 0.777462 | 3 | 0 | 3 |
| ADH1A | FLT3LG | 0.762579 | 3 | 0 | 3 |
| ADH1A | ANG | 0.727006 | 3 | 0 | 3 |
| ADH1A | GH2 | 0.715612 | 4 | 0 | 3 |
| ADH1A | CNR1 | 0.703399 | 3 | 0 | 3 |
| ADH1A | CNTF | 0.703188 | 3 | 0 | 3 |
| ADH1A | CXCR5 | 0.696611 | 3 | 0 | 3 |
| ADH1A | RAB23 | 0.692854 | 3 | 0 | 3 |
| ADH1A | CACNA2D4 | 0.688411 | 3 | 0 | 3 |
| ADH1A | PPARA | 0.686722 | 3 | 0 | 3 |
| ADH1A | KCNK2 | 0.686588 | 4 | 0 | 3 |
| ADH1A | ZNF257 | 0.669546 | 3 | 0 | 3 |
| ADH1A | ATRNL1 | 0.667485 | 3 | 0 | 3 |
| ADH1A | BTN1A1 | 0.660005 | 4 | 0 | 4 |
| ADH1A | CLCN4 | 0.656716 | 3 | 0 | 3 |
| ADH1A | ZNF717 | 0.656687 | 4 | 0 | 3 |
| ADH1A | ATXN3L | 0.653483 | 6 | 0 | 5 |
| ADH1A | OLIG2 | 0.65251 | 3 | 0 | 3 |
| ADH1A | TEX15 | 0.648328 | 4 | 0 | 4 |
| ADH1A | CASR | 0.646542 | 3 | 0 | 3 |
For details and further investigation, click here