| Full name: alcohol dehydrogenase 4 (class II), pi polypeptide | Alias Symbol: ADH-2 | ||
| Type: protein-coding gene | Cytoband: 4q23 | ||
| Entrez ID: 127 | HGNC ID: HGNC:252 | Ensembl Gene: ENSG00000198099 | OMIM ID: 103740 |
| Drug and gene relationship at DGIdb | |||
Expression of ADH4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADH4 | 127 | 223781_x_at | 0.0132 | 0.9797 | |
| GSE26886 | ADH4 | 127 | 223781_x_at | 0.1171 | 0.4146 | |
| GSE45670 | ADH4 | 127 | 223781_x_at | -0.2203 | 0.2064 | |
| GSE63941 | ADH4 | 127 | 223781_x_at | -0.0882 | 0.5755 | |
| GSE77861 | ADH4 | 127 | 223781_x_at | 0.0432 | 0.7203 | |
| GSE97050 | ADH4 | 127 | A_23_P30098 | -0.4862 | 0.1889 | |
| SRP064894 | ADH4 | 127 | RNAseq | -1.4239 | 0.0000 | |
| SRP133303 | ADH4 | 127 | RNAseq | -0.0973 | 0.4813 | |
| SRP159526 | ADH4 | 127 | RNAseq | 0.0285 | 0.9558 | |
| SRP193095 | ADH4 | 127 | RNAseq | 0.0216 | 0.9056 | |
| SRP219564 | ADH4 | 127 | RNAseq | -0.0917 | 0.8711 | |
| TCGA | ADH4 | 127 | RNAseq | -3.0544 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 2.
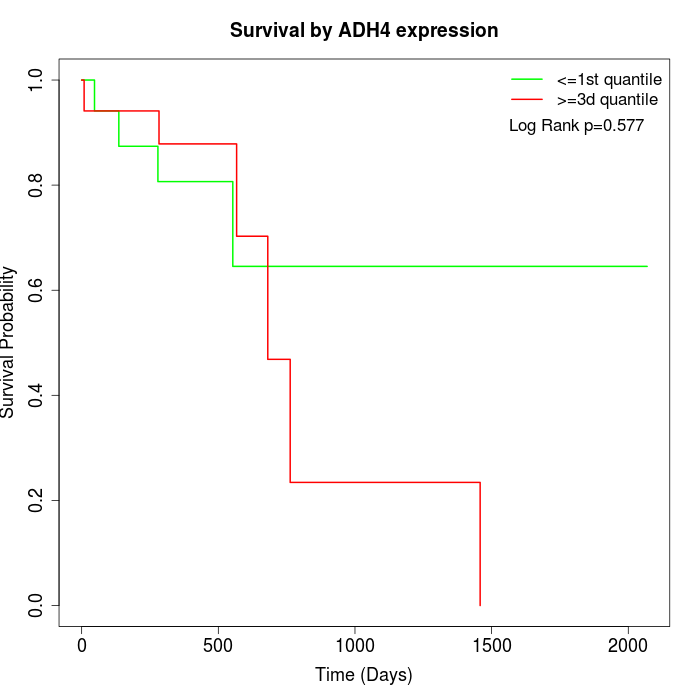
Survival by ADH4 expression:
Note: Click image to view full size file.
Copy number change of ADH4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADH4 | 127 | 0 | 13 | 17 | |
| GSE20123 | ADH4 | 127 | 0 | 13 | 17 | |
| GSE43470 | ADH4 | 127 | 0 | 14 | 29 | |
| GSE46452 | ADH4 | 127 | 1 | 36 | 22 | |
| GSE47630 | ADH4 | 127 | 0 | 20 | 20 | |
| GSE54993 | ADH4 | 127 | 9 | 0 | 61 | |
| GSE54994 | ADH4 | 127 | 1 | 11 | 41 | |
| GSE60625 | ADH4 | 127 | 0 | 3 | 8 | |
| GSE74703 | ADH4 | 127 | 0 | 12 | 24 | |
| GSE74704 | ADH4 | 127 | 0 | 6 | 14 | |
| TCGA | ADH4 | 127 | 8 | 41 | 47 |
Total number of gains: 19; Total number of losses: 169; Total Number of normals: 300.
Somatic mutations of ADH4:
Generating mutation plots.
Highly correlated genes for ADH4:
Showing top 20/62 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADH4 | DDX59 | 0.757908 | 4 | 0 | 4 |
| ADH4 | TNRC6C-AS1 | 0.754768 | 3 | 0 | 3 |
| ADH4 | C9orf64 | 0.737423 | 4 | 0 | 4 |
| ADH4 | HAUS2 | 0.735712 | 4 | 0 | 4 |
| ADH4 | FUT8-AS1 | 0.731308 | 3 | 0 | 3 |
| ADH4 | MZT2B | 0.721568 | 5 | 0 | 5 |
| ADH4 | RAMP2-AS1 | 0.721109 | 3 | 0 | 3 |
| ADH4 | NLN | 0.714125 | 4 | 0 | 4 |
| ADH4 | DIP2A | 0.706041 | 5 | 0 | 4 |
| ADH4 | CMBL | 0.696447 | 4 | 0 | 4 |
| ADH4 | CHD8 | 0.695634 | 3 | 0 | 3 |
| ADH4 | AVIL | 0.691464 | 4 | 0 | 4 |
| ADH4 | ZNF721 | 0.690002 | 5 | 0 | 4 |
| ADH4 | NKTR | 0.689404 | 4 | 0 | 3 |
| ADH4 | ENTPD2 | 0.685602 | 3 | 0 | 3 |
| ADH4 | MATR3 | 0.664235 | 3 | 0 | 3 |
| ADH4 | FAM161B | 0.659709 | 5 | 0 | 4 |
| ADH4 | SMCR5 | 0.658049 | 3 | 0 | 3 |
| ADH4 | LRRFIP1 | 0.657268 | 4 | 0 | 4 |
| ADH4 | SLC22A3 | 0.652691 | 4 | 0 | 3 |
For details and further investigation, click here