| Full name: Smith-Magenis syndrome chromosome region, candidate 5 | Alias Symbol: NCRNA00034 | ||
| Type: non-coding RNA | Cytoband: 17p11.2 | ||
| Entrez ID: 140771 | HGNC ID: HGNC:17918 | Ensembl Gene: ENSG00000226746 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SMCR5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMCR5 | 140771 | 1553020_at | 0.0342 | 0.9238 | |
| GSE26886 | SMCR5 | 140771 | 1553020_at | 0.1481 | 0.2727 | |
| GSE45670 | SMCR5 | 140771 | 1553020_at | 0.0544 | 0.5894 | |
| GSE53622 | SMCR5 | 140771 | 32257 | 0.4117 | 0.0011 | |
| GSE53624 | SMCR5 | 140771 | 32257 | 0.4025 | 0.0000 | |
| GSE63941 | SMCR5 | 140771 | 1553020_at | 0.0456 | 0.8058 | |
| GSE77861 | SMCR5 | 140771 | 1553020_at | -0.1398 | 0.0975 | |
| SRP133303 | SMCR5 | 140771 | RNAseq | 0.2333 | 0.1823 | |
| TCGA | SMCR5 | 140771 | RNAseq | -0.4816 | 0.1975 |
Upregulated datasets: 0; Downregulated datasets: 0.
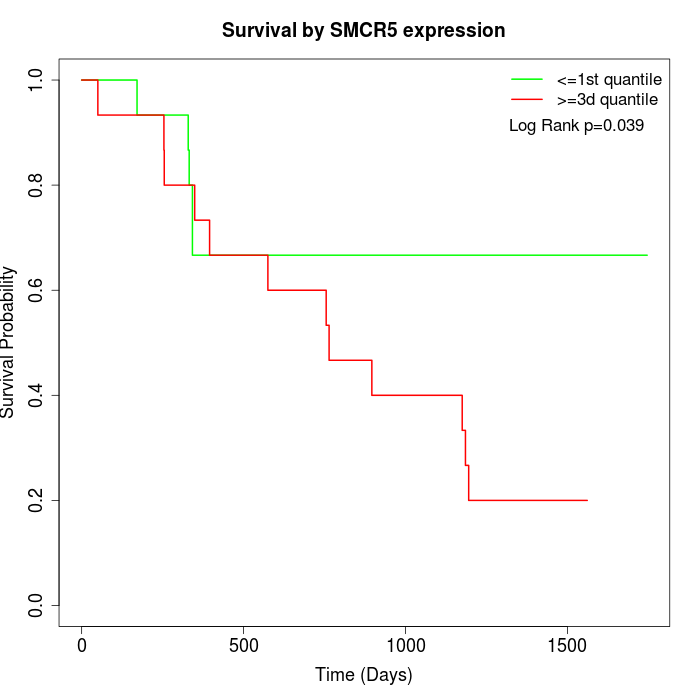
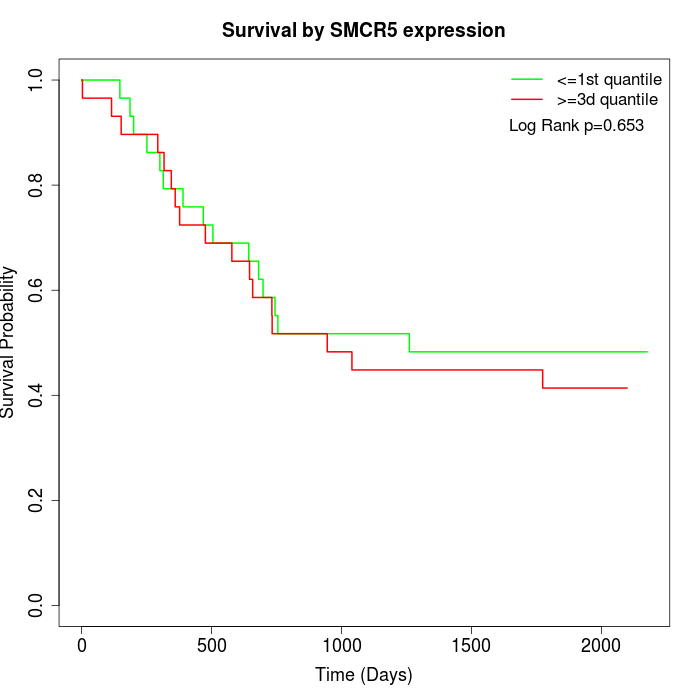
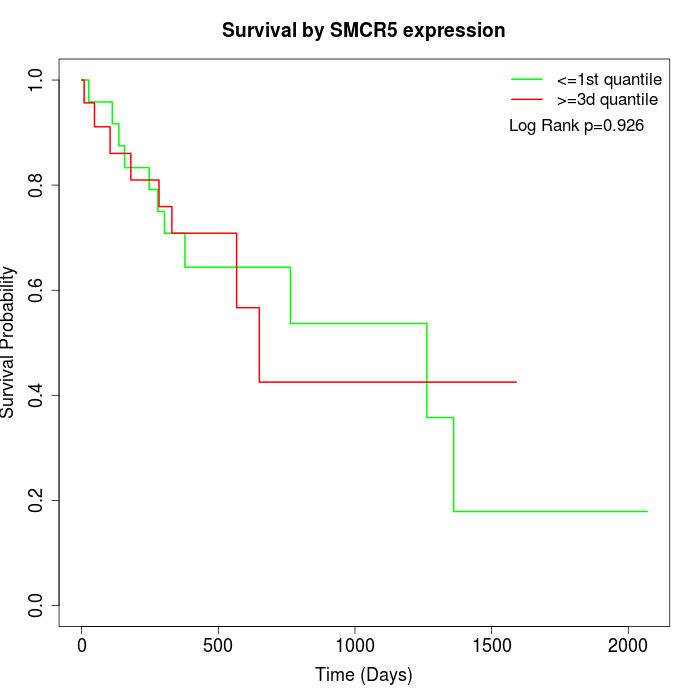
Survival by SMCR5 expression:
Note: Click image to view full size file.
Copy number change of SMCR5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMCR5 | 140771 | 3 | 3 | 24 | |
| GSE20123 | SMCR5 | 140771 | 3 | 4 | 23 | |
| GSE43470 | SMCR5 | 140771 | 1 | 5 | 37 | |
| GSE46452 | SMCR5 | 140771 | 34 | 1 | 24 | |
| GSE47630 | SMCR5 | 140771 | 7 | 1 | 32 | |
| GSE54993 | SMCR5 | 140771 | 3 | 3 | 64 | |
| GSE54994 | SMCR5 | 140771 | 6 | 6 | 41 | |
| GSE60625 | SMCR5 | 140771 | 4 | 0 | 7 | |
| GSE74703 | SMCR5 | 140771 | 1 | 2 | 33 | |
| GSE74704 | SMCR5 | 140771 | 2 | 1 | 17 | |
| TCGA | SMCR5 | 140771 | 18 | 23 | 55 |
Total number of gains: 82; Total number of losses: 49; Total Number of normals: 357.
Somatic mutations of SMCR5:
Generating mutation plots.
Highly correlated genes for SMCR5:
Showing top 20/60 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMCR5 | NEK8 | 0.738393 | 3 | 0 | 3 |
| SMCR5 | SFI1 | 0.673068 | 3 | 0 | 3 |
| SMCR5 | JRK | 0.661193 | 4 | 0 | 4 |
| SMCR5 | PNMA3 | 0.658256 | 3 | 0 | 3 |
| SMCR5 | ADH4 | 0.658049 | 3 | 0 | 3 |
| SMCR5 | XRCC2 | 0.64148 | 5 | 0 | 4 |
| SMCR5 | ABCA7 | 0.640737 | 3 | 0 | 3 |
| SMCR5 | SEPSECS-AS1 | 0.633516 | 3 | 0 | 3 |
| SMCR5 | WDR27 | 0.63144 | 4 | 0 | 4 |
| SMCR5 | LIPE-AS1 | 0.6274 | 3 | 0 | 3 |
| SMCR5 | CDAN1 | 0.623383 | 3 | 0 | 3 |
| SMCR5 | HABP2 | 0.619851 | 3 | 0 | 3 |
| SMCR5 | PDHA2 | 0.614723 | 3 | 0 | 3 |
| SMCR5 | LAMB4 | 0.61189 | 3 | 0 | 3 |
| SMCR5 | TH | 0.611442 | 3 | 0 | 3 |
| SMCR5 | ZNF444 | 0.609967 | 3 | 0 | 3 |
| SMCR5 | SND1-IT1 | 0.607473 | 4 | 0 | 3 |
| SMCR5 | ATF5 | 0.606321 | 3 | 0 | 3 |
| SMCR5 | KDM8 | 0.598211 | 3 | 0 | 3 |
| SMCR5 | RAX2 | 0.597899 | 6 | 0 | 4 |
For details and further investigation, click here