| Full name: 2-aminoethanethiol dioxygenase | Alias Symbol: FLJ14547 | ||
| Type: protein-coding gene | Cytoband: 10q21.3 | ||
| Entrez ID: 84890 | HGNC ID: HGNC:23506 | Ensembl Gene: ENSG00000181915 | OMIM ID: 611392 |
| Drug and gene relationship at DGIdb | |||
Expression of ADO:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADO | 84890 | 212502_at | 0.9212 | 0.0308 | |
| GSE20347 | ADO | 84890 | 212500_at | 0.0962 | 0.6449 | |
| GSE23400 | ADO | 84890 | 212500_at | 0.1236 | 0.0142 | |
| GSE26886 | ADO | 84890 | 212500_at | 0.3482 | 0.1409 | |
| GSE29001 | ADO | 84890 | 212502_at | 0.8360 | 0.0070 | |
| GSE38129 | ADO | 84890 | 212500_at | 0.2407 | 0.0448 | |
| GSE45670 | ADO | 84890 | 212502_at | 0.6236 | 0.0246 | |
| GSE53622 | ADO | 84890 | 3800 | 0.1627 | 0.0562 | |
| GSE53624 | ADO | 84890 | 3800 | 0.4566 | 0.0000 | |
| GSE63941 | ADO | 84890 | 212502_at | 0.4134 | 0.4327 | |
| GSE77861 | ADO | 84890 | 212502_at | 0.7538 | 0.0151 | |
| GSE97050 | ADO | 84890 | A_23_P202345 | 0.0068 | 0.9820 | |
| SRP007169 | ADO | 84890 | RNAseq | 0.5543 | 0.1899 | |
| SRP008496 | ADO | 84890 | RNAseq | 0.7612 | 0.0018 | |
| SRP064894 | ADO | 84890 | RNAseq | 0.5768 | 0.0000 | |
| SRP133303 | ADO | 84890 | RNAseq | 0.7660 | 0.0000 | |
| SRP159526 | ADO | 84890 | RNAseq | 0.4988 | 0.0157 | |
| SRP193095 | ADO | 84890 | RNAseq | 0.2632 | 0.0085 | |
| SRP219564 | ADO | 84890 | RNAseq | 0.1406 | 0.6487 | |
| TCGA | ADO | 84890 | RNAseq | 0.0993 | 0.0565 |
Upregulated datasets: 0; Downregulated datasets: 0.
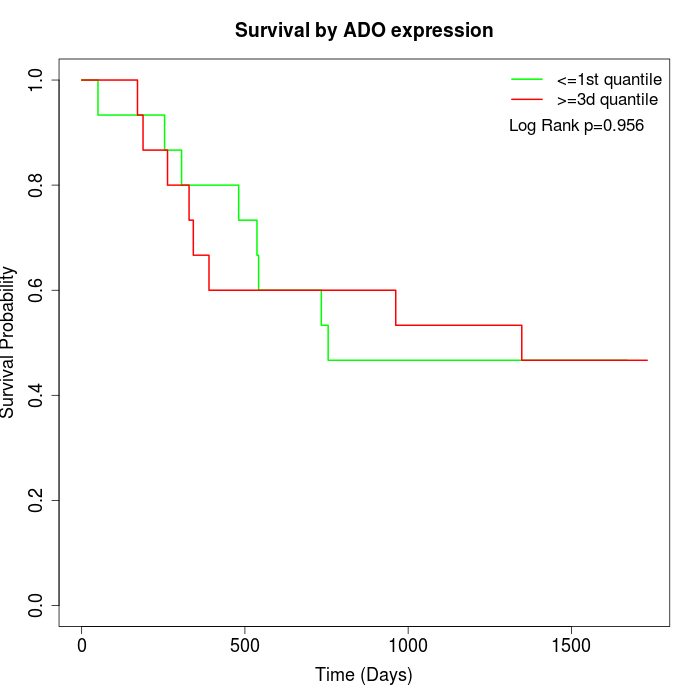
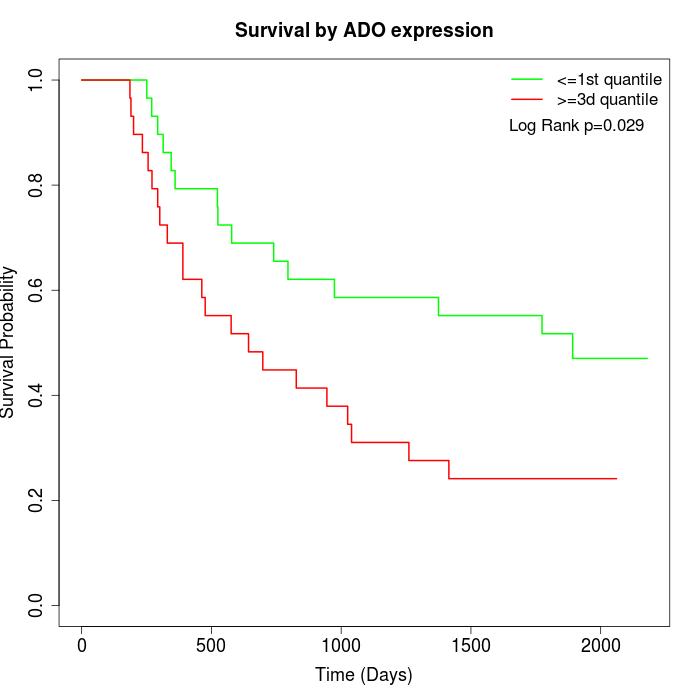
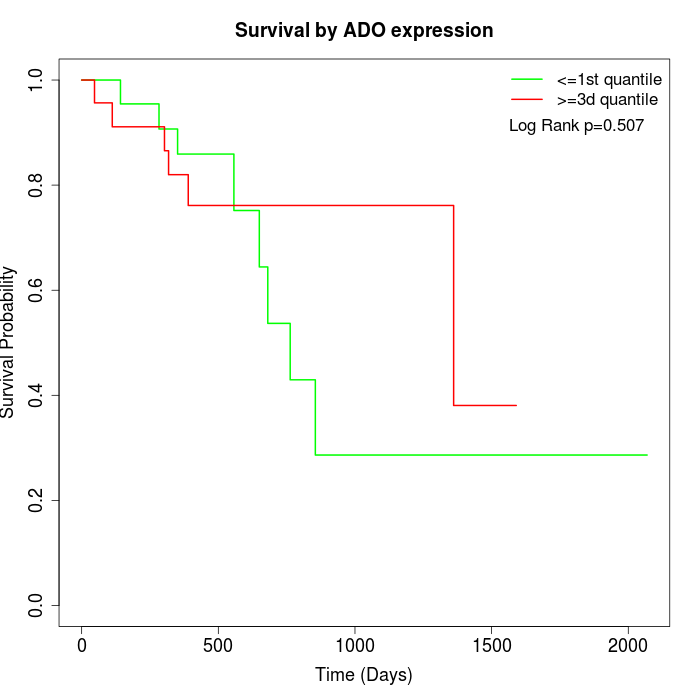
Survival by ADO expression:
Note: Click image to view full size file.
Copy number change of ADO:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADO | 84890 | 2 | 7 | 21 | |
| GSE20123 | ADO | 84890 | 2 | 7 | 21 | |
| GSE43470 | ADO | 84890 | 2 | 5 | 36 | |
| GSE46452 | ADO | 84890 | 0 | 11 | 48 | |
| GSE47630 | ADO | 84890 | 4 | 12 | 24 | |
| GSE54993 | ADO | 84890 | 9 | 0 | 61 | |
| GSE54994 | ADO | 84890 | 4 | 9 | 40 | |
| GSE60625 | ADO | 84890 | 0 | 0 | 11 | |
| GSE74703 | ADO | 84890 | 2 | 3 | 31 | |
| GSE74704 | ADO | 84890 | 1 | 5 | 14 | |
| TCGA | ADO | 84890 | 12 | 22 | 62 |
Total number of gains: 38; Total number of losses: 81; Total Number of normals: 369.
Somatic mutations of ADO:
Generating mutation plots.
Highly correlated genes for ADO:
Showing top 20/1799 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADO | DUS3L | 0.833701 | 3 | 0 | 3 |
| ADO | ORAI1 | 0.803813 | 3 | 0 | 3 |
| ADO | NAPB | 0.772637 | 3 | 0 | 3 |
| ADO | DNAJC19 | 0.76566 | 3 | 0 | 3 |
| ADO | TBC1D8 | 0.757844 | 3 | 0 | 3 |
| ADO | FAM160B1 | 0.750971 | 3 | 0 | 3 |
| ADO | MMP15 | 0.750396 | 3 | 0 | 3 |
| ADO | VMA21 | 0.748532 | 4 | 0 | 4 |
| ADO | ATG4C | 0.746348 | 3 | 0 | 3 |
| ADO | PRRX2 | 0.741407 | 3 | 0 | 3 |
| ADO | TRIM59 | 0.74104 | 3 | 0 | 3 |
| ADO | SLC2A3 | 0.740831 | 3 | 0 | 3 |
| ADO | SMARCAD1 | 0.739503 | 4 | 0 | 4 |
| ADO | CMTM4 | 0.738927 | 3 | 0 | 3 |
| ADO | DDB1 | 0.738491 | 3 | 0 | 3 |
| ADO | TMEM138 | 0.738297 | 5 | 0 | 5 |
| ADO | HPRT1 | 0.735625 | 4 | 0 | 4 |
| ADO | PLAGL2 | 0.735279 | 4 | 0 | 3 |
| ADO | FOXJ2 | 0.734562 | 5 | 0 | 5 |
| ADO | BTBD2 | 0.731641 | 3 | 0 | 3 |
For details and further investigation, click here