| Full name: SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 | Alias Symbol: ETL1|DKFZP762K2015|KIAA1122|DKFZp762K2015 | ||
| Type: protein-coding gene | Cytoband: 4q22.3 | ||
| Entrez ID: 56916 | HGNC ID: HGNC:18398 | Ensembl Gene: ENSG00000163104 | OMIM ID: 612761 |
| Drug and gene relationship at DGIdb | |||
Expression of SMARCAD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMARCAD1 | 56916 | 223197_s_at | 0.4197 | 0.0779 | |
| GSE26886 | SMARCAD1 | 56916 | 223197_s_at | -0.2580 | 0.2531 | |
| GSE45670 | SMARCAD1 | 56916 | 223197_s_at | 0.1204 | 0.5052 | |
| GSE53622 | SMARCAD1 | 56916 | 72880 | 0.2909 | 0.0002 | |
| GSE53624 | SMARCAD1 | 56916 | 21993 | 0.0967 | 0.2568 | |
| GSE63941 | SMARCAD1 | 56916 | 223197_s_at | -0.5262 | 0.2801 | |
| GSE77861 | SMARCAD1 | 56916 | 223197_s_at | 0.0353 | 0.9172 | |
| GSE97050 | SMARCAD1 | 56916 | A_33_P3351474 | 0.2615 | 0.2687 | |
| SRP007169 | SMARCAD1 | 56916 | RNAseq | 0.0711 | 0.8632 | |
| SRP008496 | SMARCAD1 | 56916 | RNAseq | 0.2404 | 0.2952 | |
| SRP064894 | SMARCAD1 | 56916 | RNAseq | -0.0536 | 0.8364 | |
| SRP133303 | SMARCAD1 | 56916 | RNAseq | 0.1582 | 0.3400 | |
| SRP159526 | SMARCAD1 | 56916 | RNAseq | 0.1040 | 0.7497 | |
| SRP193095 | SMARCAD1 | 56916 | RNAseq | -0.1223 | 0.2760 | |
| SRP219564 | SMARCAD1 | 56916 | RNAseq | -0.2810 | 0.4072 | |
| TCGA | SMARCAD1 | 56916 | RNAseq | 0.0224 | 0.6529 |
Upregulated datasets: 0; Downregulated datasets: 0.
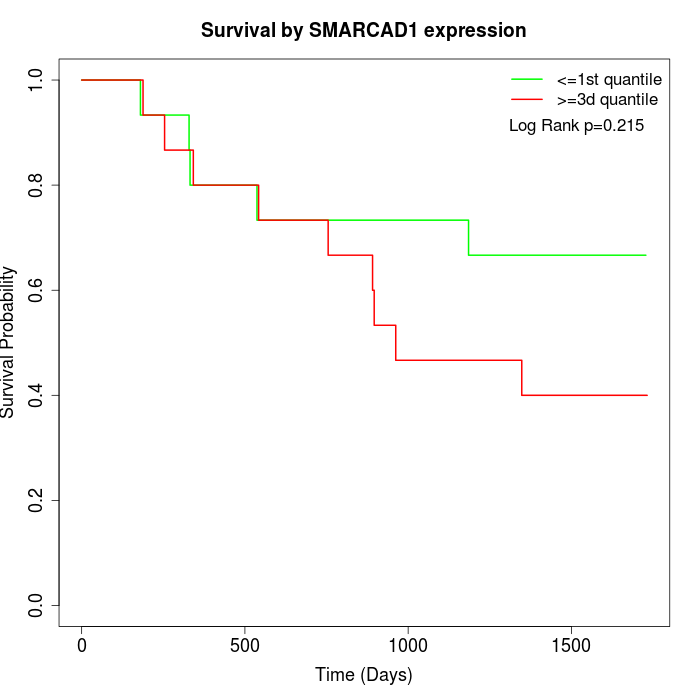
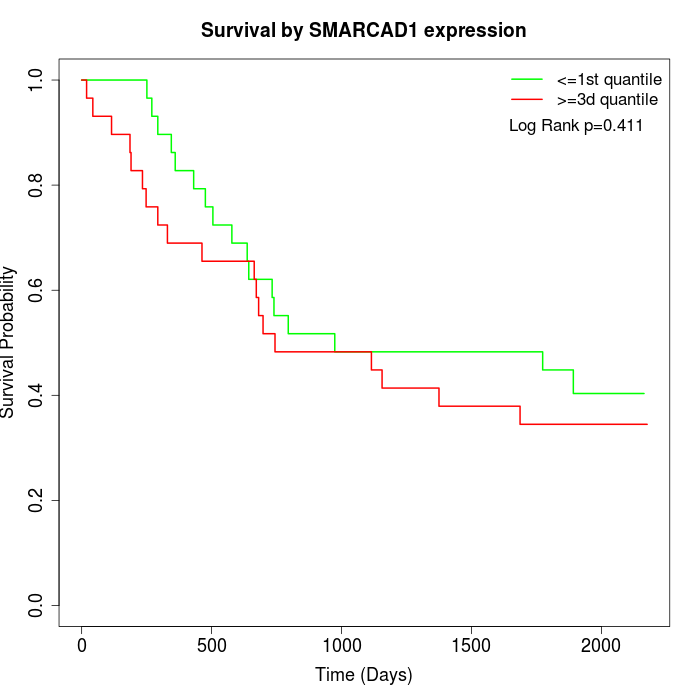
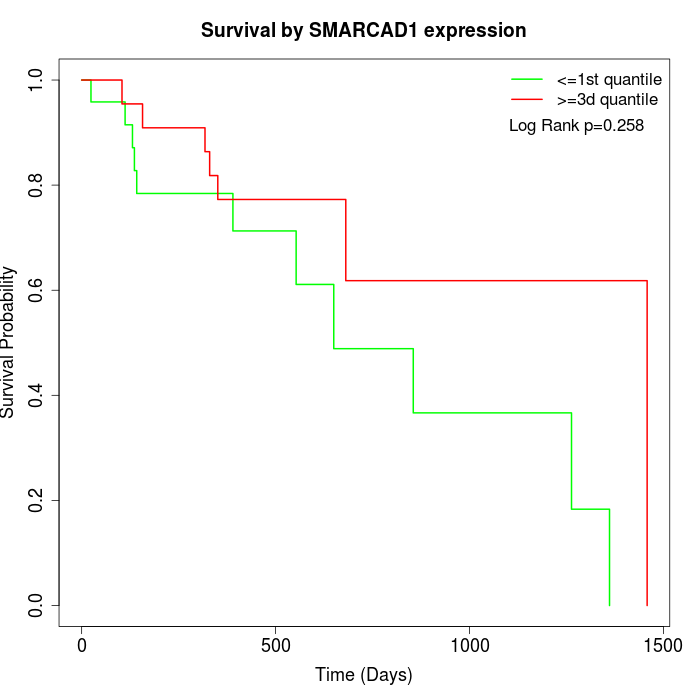
Survival by SMARCAD1 expression:
Note: Click image to view full size file.
Copy number change of SMARCAD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMARCAD1 | 56916 | 0 | 12 | 18 | |
| GSE20123 | SMARCAD1 | 56916 | 0 | 12 | 18 | |
| GSE43470 | SMARCAD1 | 56916 | 0 | 16 | 27 | |
| GSE46452 | SMARCAD1 | 56916 | 1 | 36 | 22 | |
| GSE47630 | SMARCAD1 | 56916 | 0 | 20 | 20 | |
| GSE54993 | SMARCAD1 | 56916 | 8 | 0 | 62 | |
| GSE54994 | SMARCAD1 | 56916 | 1 | 11 | 41 | |
| GSE60625 | SMARCAD1 | 56916 | 0 | 3 | 8 | |
| GSE74703 | SMARCAD1 | 56916 | 0 | 14 | 22 | |
| GSE74704 | SMARCAD1 | 56916 | 0 | 6 | 14 | |
| TCGA | SMARCAD1 | 56916 | 8 | 40 | 48 |
Total number of gains: 18; Total number of losses: 170; Total Number of normals: 300.
Somatic mutations of SMARCAD1:
Generating mutation plots.
Highly correlated genes for SMARCAD1:
Showing top 20/404 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMARCAD1 | NOLC1 | 0.822154 | 3 | 0 | 3 |
| SMARCAD1 | DNASE2 | 0.810068 | 3 | 0 | 3 |
| SMARCAD1 | ZC3HC1 | 0.799477 | 3 | 0 | 3 |
| SMARCAD1 | ABCF1 | 0.796333 | 3 | 0 | 3 |
| SMARCAD1 | SBNO1 | 0.774671 | 3 | 0 | 3 |
| SMARCAD1 | GMFB | 0.75779 | 3 | 0 | 3 |
| SMARCAD1 | DNAJC9 | 0.752788 | 4 | 0 | 3 |
| SMARCAD1 | LETMD1 | 0.749302 | 3 | 0 | 3 |
| SMARCAD1 | C2CD2 | 0.748459 | 3 | 0 | 3 |
| SMARCAD1 | SPG21 | 0.747657 | 3 | 0 | 3 |
| SMARCAD1 | SNRPD2 | 0.747378 | 3 | 0 | 3 |
| SMARCAD1 | STK4 | 0.747161 | 3 | 0 | 3 |
| SMARCAD1 | CCNT1 | 0.743615 | 3 | 0 | 3 |
| SMARCAD1 | ASCC1 | 0.740598 | 3 | 0 | 3 |
| SMARCAD1 | JTB | 0.739729 | 3 | 0 | 3 |
| SMARCAD1 | ADO | 0.739503 | 4 | 0 | 4 |
| SMARCAD1 | CCDC15 | 0.738735 | 3 | 0 | 3 |
| SMARCAD1 | ARL6 | 0.737888 | 3 | 0 | 3 |
| SMARCAD1 | CHAMP1 | 0.736826 | 3 | 0 | 3 |
| SMARCAD1 | ALDH18A1 | 0.735709 | 4 | 0 | 4 |
For details and further investigation, click here