| Full name: amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p21.2 | ||
| Entrez ID: 178 | HGNC ID: HGNC:321 | Ensembl Gene: ENSG00000162688 | OMIM ID: 610860 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of AGL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AGL | 178 | 203566_s_at | -0.0957 | 0.9138 | |
| GSE20347 | AGL | 178 | 203566_s_at | -0.4178 | 0.0371 | |
| GSE23400 | AGL | 178 | 203566_s_at | -0.0888 | 0.5540 | |
| GSE26886 | AGL | 178 | 203566_s_at | -0.4615 | 0.1282 | |
| GSE29001 | AGL | 178 | 203566_s_at | 0.1270 | 0.6847 | |
| GSE38129 | AGL | 178 | 203566_s_at | -0.5212 | 0.0006 | |
| GSE45670 | AGL | 178 | 203566_s_at | -0.3878 | 0.0550 | |
| GSE53622 | AGL | 178 | 94524 | -0.7496 | 0.0000 | |
| GSE53624 | AGL | 178 | 94524 | -0.4629 | 0.0000 | |
| GSE63941 | AGL | 178 | 203566_s_at | 0.1675 | 0.7976 | |
| GSE77861 | AGL | 178 | 203566_s_at | -0.6000 | 0.1522 | |
| GSE97050 | AGL | 178 | A_23_P200298 | -1.0552 | 0.0844 | |
| SRP007169 | AGL | 178 | RNAseq | 1.0338 | 0.0419 | |
| SRP008496 | AGL | 178 | RNAseq | 0.9226 | 0.0007 | |
| SRP064894 | AGL | 178 | RNAseq | -0.1097 | 0.6594 | |
| SRP133303 | AGL | 178 | RNAseq | -0.1550 | 0.4790 | |
| SRP159526 | AGL | 178 | RNAseq | -0.9556 | 0.0022 | |
| SRP193095 | AGL | 178 | RNAseq | -0.2296 | 0.0520 | |
| SRP219564 | AGL | 178 | RNAseq | -0.6780 | 0.2297 | |
| TCGA | AGL | 178 | RNAseq | -0.1328 | 0.0331 |
Upregulated datasets: 1; Downregulated datasets: 0.
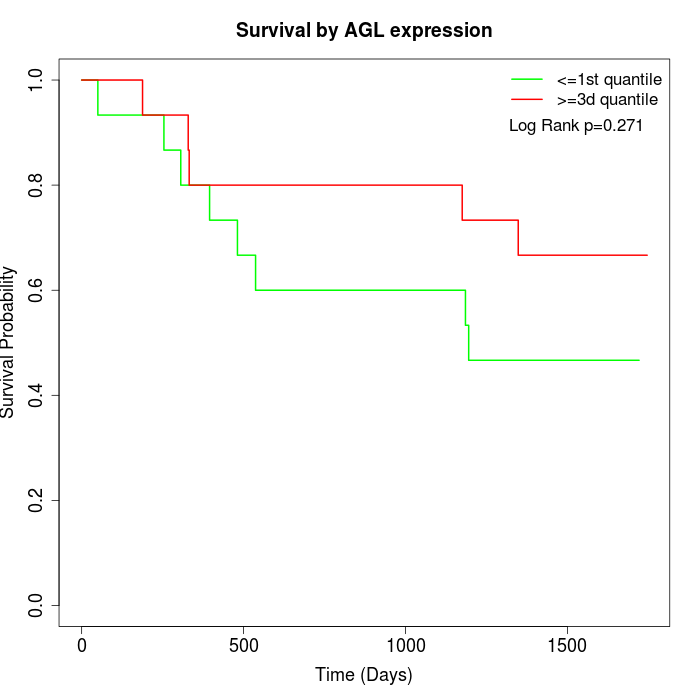
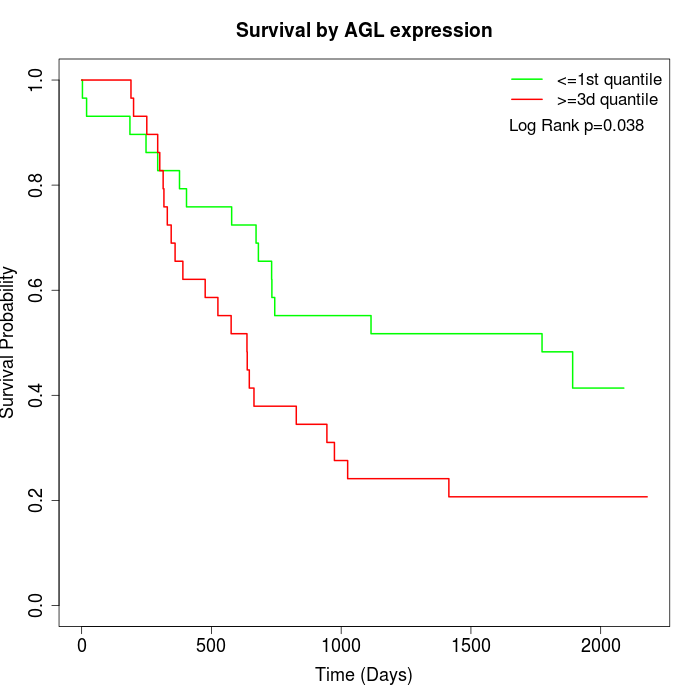
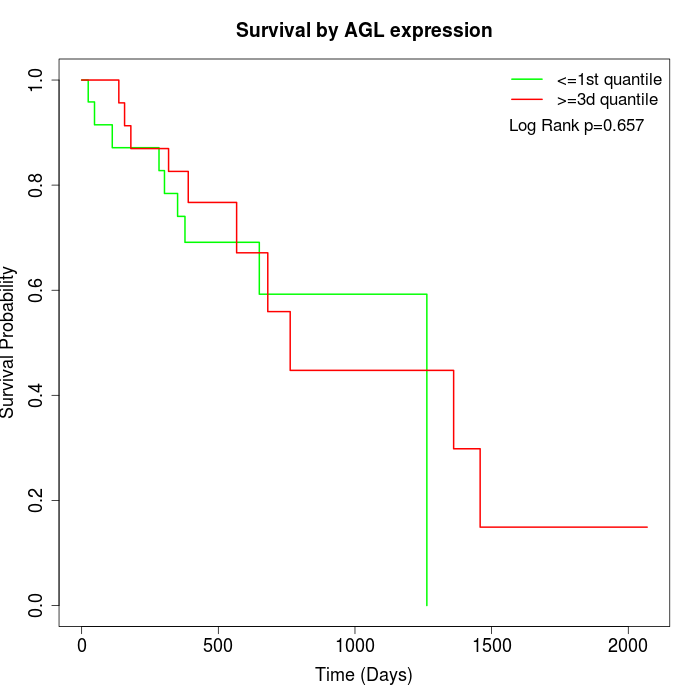
Survival by AGL expression:
Note: Click image to view full size file.
Copy number change of AGL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AGL | 178 | 0 | 10 | 20 | |
| GSE20123 | AGL | 178 | 0 | 10 | 20 | |
| GSE43470 | AGL | 178 | 1 | 6 | 36 | |
| GSE46452 | AGL | 178 | 1 | 1 | 57 | |
| GSE47630 | AGL | 178 | 9 | 5 | 26 | |
| GSE54993 | AGL | 178 | 0 | 1 | 69 | |
| GSE54994 | AGL | 178 | 6 | 3 | 44 | |
| GSE60625 | AGL | 178 | 0 | 0 | 11 | |
| GSE74703 | AGL | 178 | 0 | 5 | 31 | |
| GSE74704 | AGL | 178 | 0 | 6 | 14 | |
| TCGA | AGL | 178 | 8 | 24 | 64 |
Total number of gains: 25; Total number of losses: 71; Total Number of normals: 392.
Somatic mutations of AGL:
Generating mutation plots.
Highly correlated genes for AGL:
Showing top 20/589 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AGL | PTPRG | 0.781036 | 3 | 0 | 3 |
| AGL | NUDT21 | 0.778886 | 3 | 0 | 3 |
| AGL | NTAN1 | 0.752087 | 3 | 0 | 3 |
| AGL | C2CD2 | 0.744079 | 3 | 0 | 3 |
| AGL | ZFAND6 | 0.741111 | 3 | 0 | 3 |
| AGL | AP3M1 | 0.739546 | 3 | 0 | 3 |
| AGL | ROCK2 | 0.733847 | 3 | 0 | 3 |
| AGL | BBS9 | 0.727905 | 4 | 0 | 3 |
| AGL | TCEANC2 | 0.726418 | 3 | 0 | 3 |
| AGL | MIER3 | 0.720238 | 4 | 0 | 4 |
| AGL | RAD50 | 0.716738 | 4 | 0 | 4 |
| AGL | BLOC1S2 | 0.712871 | 3 | 0 | 3 |
| AGL | PIGK | 0.710832 | 4 | 0 | 3 |
| AGL | ZFX | 0.709893 | 3 | 0 | 3 |
| AGL | ANAPC16 | 0.707647 | 4 | 0 | 4 |
| AGL | GNAT2 | 0.706781 | 3 | 0 | 3 |
| AGL | FBXL3 | 0.700625 | 4 | 0 | 4 |
| AGL | PTCD2 | 0.698446 | 4 | 0 | 4 |
| AGL | C5orf24 | 0.696594 | 4 | 0 | 3 |
| AGL | BIRC2 | 0.691845 | 3 | 0 | 3 |
For details and further investigation, click here