| Full name: RAD50 double strand break repair protein | Alias Symbol: hRad50|RAD50-2 | ||
| Type: protein-coding gene | Cytoband: 5q31.1 | ||
| Entrez ID: 10111 | HGNC ID: HGNC:9816 | Ensembl Gene: ENSG00000113522 | OMIM ID: 604040 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RAD50:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAD50 | 10111 | 208393_s_at | -0.2306 | 0.6336 | |
| GSE20347 | RAD50 | 10111 | 208393_s_at | -0.1462 | 0.4840 | |
| GSE23400 | RAD50 | 10111 | 208393_s_at | 0.2689 | 0.0002 | |
| GSE26886 | RAD50 | 10111 | 208393_s_at | -0.0851 | 0.7988 | |
| GSE29001 | RAD50 | 10111 | 208393_s_at | 0.0735 | 0.8411 | |
| GSE38129 | RAD50 | 10111 | 208393_s_at | 0.1185 | 0.4333 | |
| GSE45670 | RAD50 | 10111 | 208393_s_at | 0.3121 | 0.0440 | |
| GSE53622 | RAD50 | 10111 | 118183 | -0.4460 | 0.0000 | |
| GSE53624 | RAD50 | 10111 | 118183 | -0.0871 | 0.2875 | |
| GSE63941 | RAD50 | 10111 | 208393_s_at | -0.0124 | 0.9877 | |
| GSE77861 | RAD50 | 10111 | 208393_s_at | 0.0031 | 0.9943 | |
| GSE97050 | RAD50 | 10111 | A_23_P250404 | -0.3440 | 0.2204 | |
| SRP007169 | RAD50 | 10111 | RNAseq | -0.6838 | 0.0345 | |
| SRP008496 | RAD50 | 10111 | RNAseq | -0.3332 | 0.1452 | |
| SRP064894 | RAD50 | 10111 | RNAseq | -0.2580 | 0.1765 | |
| SRP133303 | RAD50 | 10111 | RNAseq | 0.0689 | 0.6874 | |
| SRP159526 | RAD50 | 10111 | RNAseq | -0.2753 | 0.5291 | |
| SRP193095 | RAD50 | 10111 | RNAseq | -0.2049 | 0.0368 | |
| SRP219564 | RAD50 | 10111 | RNAseq | -0.4925 | 0.0306 | |
| TCGA | RAD50 | 10111 | RNAseq | -0.1266 | 0.0075 |
Upregulated datasets: 0; Downregulated datasets: 0.
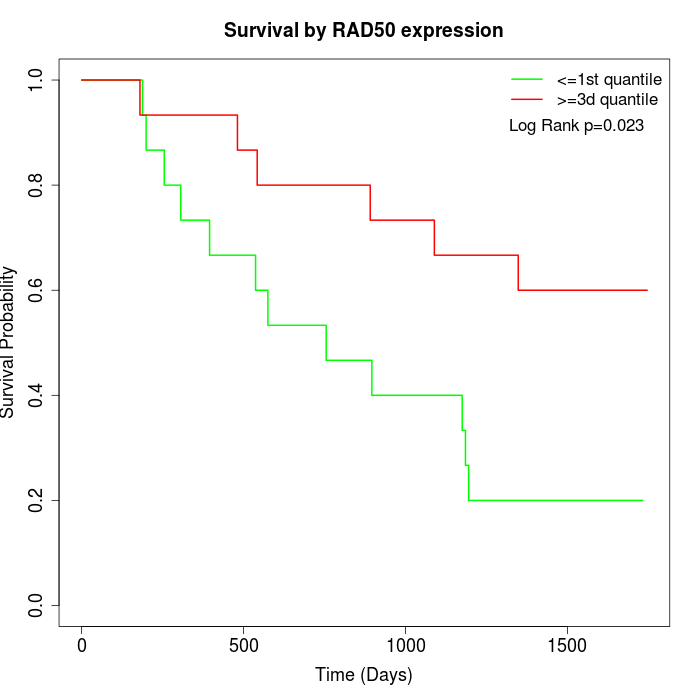
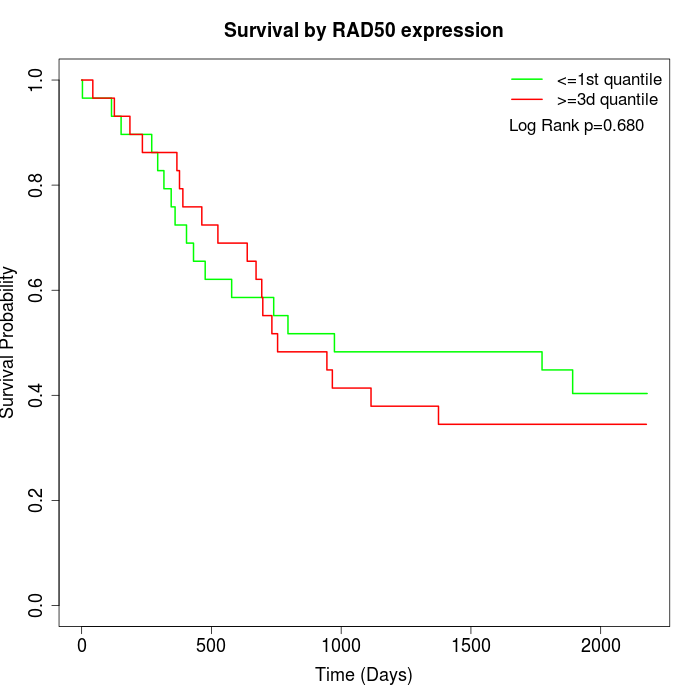
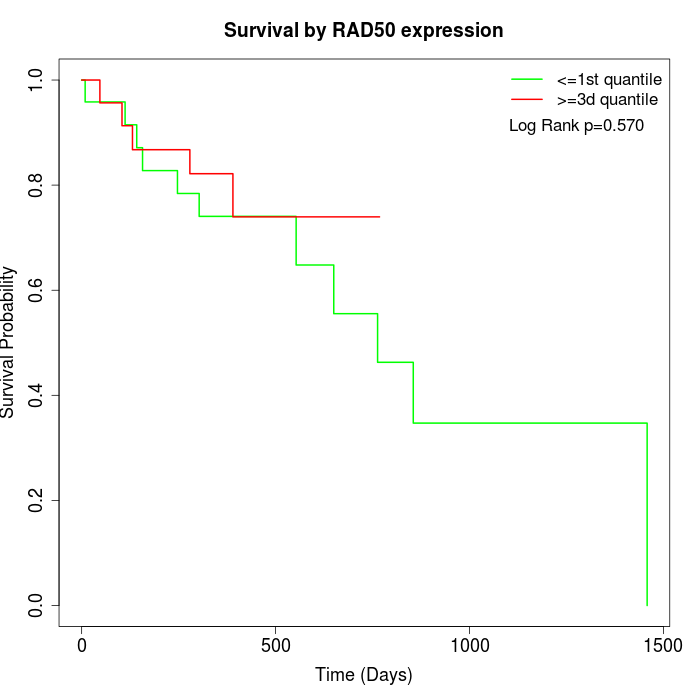
Survival by RAD50 expression:
Note: Click image to view full size file.
Copy number change of RAD50:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAD50 | 10111 | 1 | 11 | 18 | |
| GSE20123 | RAD50 | 10111 | 2 | 11 | 17 | |
| GSE43470 | RAD50 | 10111 | 2 | 9 | 32 | |
| GSE46452 | RAD50 | 10111 | 0 | 27 | 32 | |
| GSE47630 | RAD50 | 10111 | 0 | 21 | 19 | |
| GSE54993 | RAD50 | 10111 | 9 | 1 | 60 | |
| GSE54994 | RAD50 | 10111 | 2 | 14 | 37 | |
| GSE60625 | RAD50 | 10111 | 0 | 0 | 11 | |
| GSE74703 | RAD50 | 10111 | 2 | 6 | 28 | |
| GSE74704 | RAD50 | 10111 | 1 | 5 | 14 | |
| TCGA | RAD50 | 10111 | 3 | 39 | 54 |
Total number of gains: 22; Total number of losses: 144; Total Number of normals: 322.
Somatic mutations of RAD50:
Generating mutation plots.
Highly correlated genes for RAD50:
Showing top 20/410 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAD50 | LRRC28 | 0.753007 | 3 | 0 | 3 |
| RAD50 | TM4SF18 | 0.746568 | 3 | 0 | 3 |
| RAD50 | PTPRG | 0.746051 | 3 | 0 | 3 |
| RAD50 | ZNF354A | 0.733438 | 3 | 0 | 3 |
| RAD50 | TINF2 | 0.72665 | 3 | 0 | 3 |
| RAD50 | SEC22C | 0.72394 | 3 | 0 | 3 |
| RAD50 | UFL1 | 0.718475 | 3 | 0 | 3 |
| RAD50 | AGL | 0.716738 | 4 | 0 | 4 |
| RAD50 | COG6 | 0.716345 | 3 | 0 | 3 |
| RAD50 | FNIP1 | 0.711368 | 4 | 0 | 3 |
| RAD50 | GNG12 | 0.710296 | 4 | 0 | 3 |
| RAD50 | C16orf87 | 0.709452 | 4 | 0 | 4 |
| RAD50 | TTC8 | 0.706624 | 3 | 0 | 3 |
| RAD50 | NSFL1C | 0.705298 | 3 | 0 | 3 |
| RAD50 | RELL2 | 0.703166 | 3 | 0 | 3 |
| RAD50 | ZNF319 | 0.702722 | 3 | 0 | 3 |
| RAD50 | EHBP1 | 0.701044 | 3 | 0 | 3 |
| RAD50 | DUSP6 | 0.69453 | 3 | 0 | 3 |
| RAD50 | SWAP70 | 0.693426 | 3 | 0 | 3 |
| RAD50 | LARS2 | 0.690713 | 3 | 0 | 3 |
For details and further investigation, click here