| Full name: adaptor related protein complex 1 subunit sigma 1 | Alias Symbol: AP19|SIGMA1A|WUGSC:H_DJ0747G18.2 | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 1174 | HGNC ID: HGNC:559 | Ensembl Gene: ENSG00000106367 | OMIM ID: 603531 |
| Drug and gene relationship at DGIdb | |||
Expression of AP1S1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AP1S1 | 1174 | 209635_at | 0.5984 | 0.0936 | |
| GSE20347 | AP1S1 | 1174 | 209635_at | 0.2026 | 0.1629 | |
| GSE23400 | AP1S1 | 1174 | 209635_at | 0.2828 | 0.0000 | |
| GSE26886 | AP1S1 | 1174 | 209635_at | 0.5827 | 0.0004 | |
| GSE29001 | AP1S1 | 1174 | 209635_at | -0.0653 | 0.7896 | |
| GSE38129 | AP1S1 | 1174 | 209635_at | 0.6311 | 0.0004 | |
| GSE45670 | AP1S1 | 1174 | 209635_at | 0.5545 | 0.0005 | |
| GSE53622 | AP1S1 | 1174 | 116868 | 0.5899 | 0.0000 | |
| GSE53624 | AP1S1 | 1174 | 116868 | 0.4184 | 0.0000 | |
| GSE63941 | AP1S1 | 1174 | 209635_at | -0.3828 | 0.6919 | |
| GSE77861 | AP1S1 | 1174 | 209635_at | 0.2578 | 0.1860 | |
| GSE97050 | AP1S1 | 1174 | A_23_P157404 | 0.6348 | 0.1269 | |
| SRP007169 | AP1S1 | 1174 | RNAseq | -0.1768 | 0.6564 | |
| SRP008496 | AP1S1 | 1174 | RNAseq | 0.1266 | 0.7245 | |
| SRP064894 | AP1S1 | 1174 | RNAseq | 0.6864 | 0.0130 | |
| SRP133303 | AP1S1 | 1174 | RNAseq | 0.5233 | 0.0226 | |
| SRP159526 | AP1S1 | 1174 | RNAseq | 0.4627 | 0.0567 | |
| SRP193095 | AP1S1 | 1174 | RNAseq | 0.2687 | 0.1313 | |
| SRP219564 | AP1S1 | 1174 | RNAseq | 0.6278 | 0.0054 | |
| TCGA | AP1S1 | 1174 | RNAseq | 0.2928 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
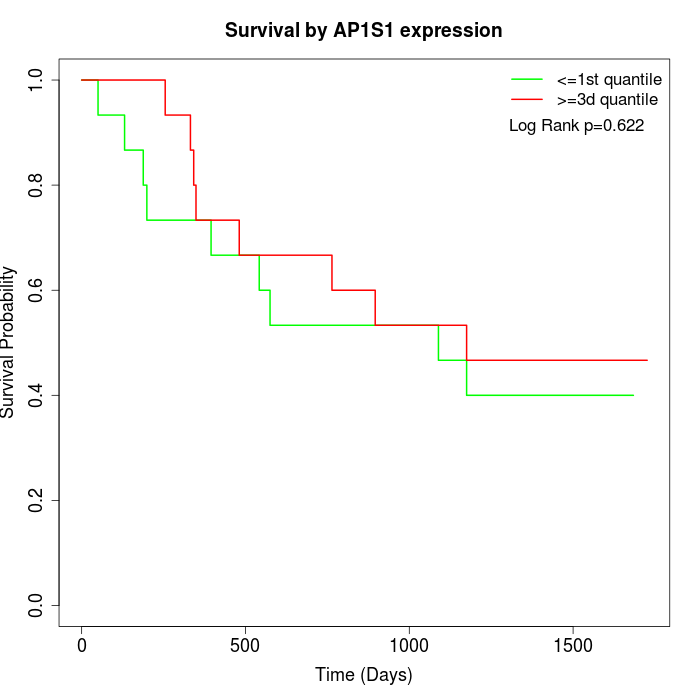
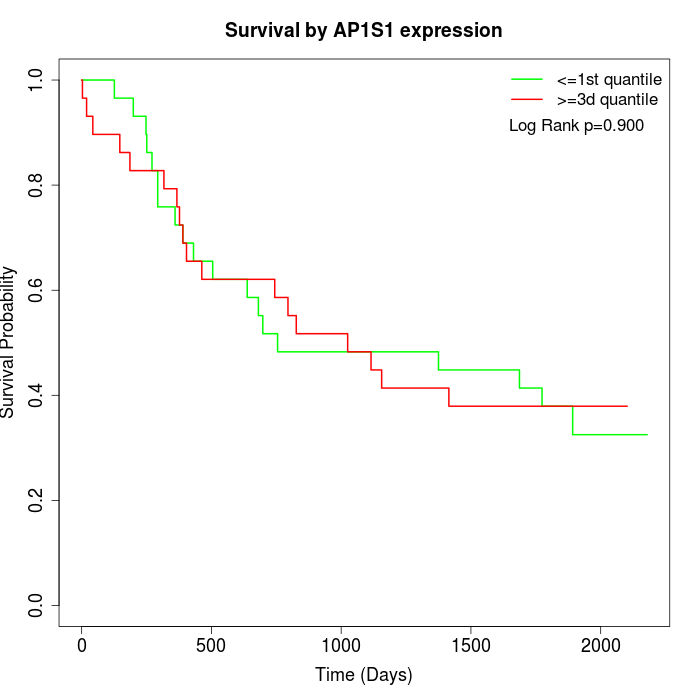
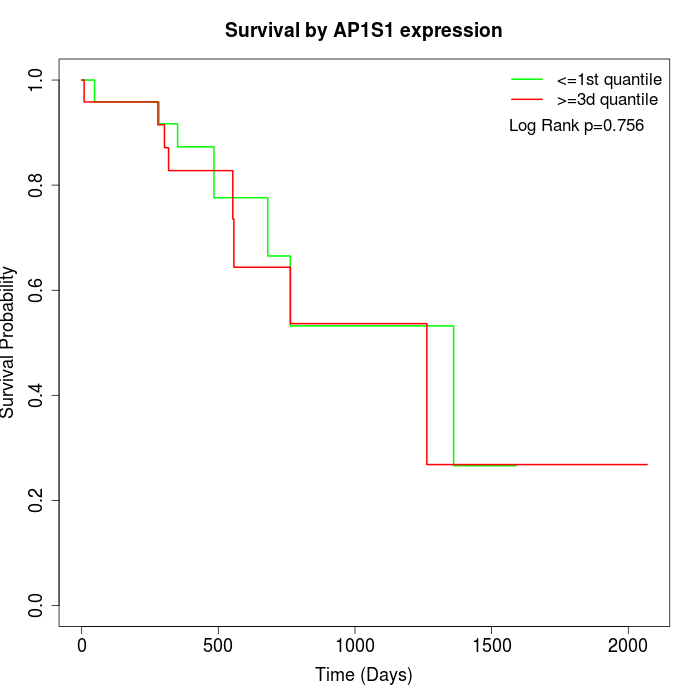
Survival by AP1S1 expression:
Note: Click image to view full size file.
Copy number change of AP1S1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AP1S1 | 1174 | 13 | 0 | 17 | |
| GSE20123 | AP1S1 | 1174 | 13 | 0 | 17 | |
| GSE43470 | AP1S1 | 1174 | 7 | 2 | 34 | |
| GSE46452 | AP1S1 | 1174 | 11 | 1 | 47 | |
| GSE47630 | AP1S1 | 1174 | 7 | 3 | 30 | |
| GSE54993 | AP1S1 | 1174 | 1 | 10 | 59 | |
| GSE54994 | AP1S1 | 1174 | 15 | 3 | 35 | |
| GSE60625 | AP1S1 | 1174 | 0 | 0 | 11 | |
| GSE74703 | AP1S1 | 1174 | 7 | 1 | 28 | |
| GSE74704 | AP1S1 | 1174 | 9 | 0 | 11 | |
| TCGA | AP1S1 | 1174 | 53 | 6 | 37 |
Total number of gains: 136; Total number of losses: 26; Total Number of normals: 326.
Somatic mutations of AP1S1:
Generating mutation plots.
Highly correlated genes for AP1S1:
Showing top 20/391 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AP1S1 | LRWD1 | 0.733799 | 3 | 0 | 3 |
| AP1S1 | HSPBAP1 | 0.706517 | 4 | 0 | 4 |
| AP1S1 | EME1 | 0.703852 | 4 | 0 | 4 |
| AP1S1 | ARHGAP23 | 0.66679 | 3 | 0 | 3 |
| AP1S1 | YRDC | 0.654536 | 4 | 0 | 3 |
| AP1S1 | TNFAIP8L1 | 0.651418 | 4 | 0 | 4 |
| AP1S1 | CLDN14 | 0.650533 | 3 | 0 | 3 |
| AP1S1 | ZNF205 | 0.637381 | 4 | 0 | 3 |
| AP1S1 | DUSP9 | 0.632234 | 4 | 0 | 3 |
| AP1S1 | WDR90 | 0.631974 | 6 | 0 | 5 |
| AP1S1 | NAV3 | 0.629084 | 4 | 0 | 4 |
| AP1S1 | WRAP73 | 0.627942 | 5 | 0 | 3 |
| AP1S1 | ZNF687 | 0.627796 | 5 | 0 | 4 |
| AP1S1 | CCDC34 | 0.626986 | 6 | 0 | 5 |
| AP1S1 | WRAP53 | 0.626413 | 6 | 0 | 4 |
| AP1S1 | CDCA5 | 0.625714 | 7 | 0 | 6 |
| AP1S1 | TMC8 | 0.622745 | 3 | 0 | 3 |
| AP1S1 | DPF1 | 0.621599 | 4 | 0 | 3 |
| AP1S1 | CHMP6 | 0.620594 | 3 | 0 | 3 |
| AP1S1 | DCBLD2 | 0.620149 | 3 | 0 | 3 |
For details and further investigation, click here