| Full name: Rho GTPase activating protein 1 | Alias Symbol: RhoGAP|p50rhoGAP|CDC42GAP|Cdc42GAP | ||
| Type: protein-coding gene | Cytoband: 11p11.2 | ||
| Entrez ID: 392 | HGNC ID: HGNC:673 | Ensembl Gene: ENSG00000175220 | OMIM ID: 602732 |
| Drug and gene relationship at DGIdb | |||
Expression of ARHGAP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGAP1 | 392 | 202117_at | -0.4019 | 0.1339 | |
| GSE20347 | ARHGAP1 | 392 | 202117_at | -0.6225 | 0.0000 | |
| GSE23400 | ARHGAP1 | 392 | 202117_at | -0.2162 | 0.0001 | |
| GSE26886 | ARHGAP1 | 392 | 202117_at | -0.3395 | 0.0664 | |
| GSE29001 | ARHGAP1 | 392 | 202117_at | -0.2965 | 0.2330 | |
| GSE38129 | ARHGAP1 | 392 | 202117_at | -0.5792 | 0.0000 | |
| GSE45670 | ARHGAP1 | 392 | 202117_at | -0.3484 | 0.0045 | |
| GSE53622 | ARHGAP1 | 392 | 20052 | -0.4815 | 0.0000 | |
| GSE53624 | ARHGAP1 | 392 | 20052 | -0.2948 | 0.0000 | |
| GSE63941 | ARHGAP1 | 392 | 202117_at | -0.7573 | 0.0647 | |
| GSE77861 | ARHGAP1 | 392 | 202117_at | -0.2193 | 0.4834 | |
| GSE97050 | ARHGAP1 | 392 | A_23_P314070 | -0.2383 | 0.2818 | |
| SRP007169 | ARHGAP1 | 392 | RNAseq | -1.2053 | 0.0030 | |
| SRP008496 | ARHGAP1 | 392 | RNAseq | -1.2117 | 0.0000 | |
| SRP064894 | ARHGAP1 | 392 | RNAseq | 0.0389 | 0.7761 | |
| SRP133303 | ARHGAP1 | 392 | RNAseq | -0.2914 | 0.1168 | |
| SRP159526 | ARHGAP1 | 392 | RNAseq | -0.8639 | 0.0008 | |
| SRP193095 | ARHGAP1 | 392 | RNAseq | -0.2067 | 0.1973 | |
| SRP219564 | ARHGAP1 | 392 | RNAseq | -0.2469 | 0.5400 | |
| TCGA | ARHGAP1 | 392 | RNAseq | -0.0464 | 0.2740 |
Upregulated datasets: 0; Downregulated datasets: 2.
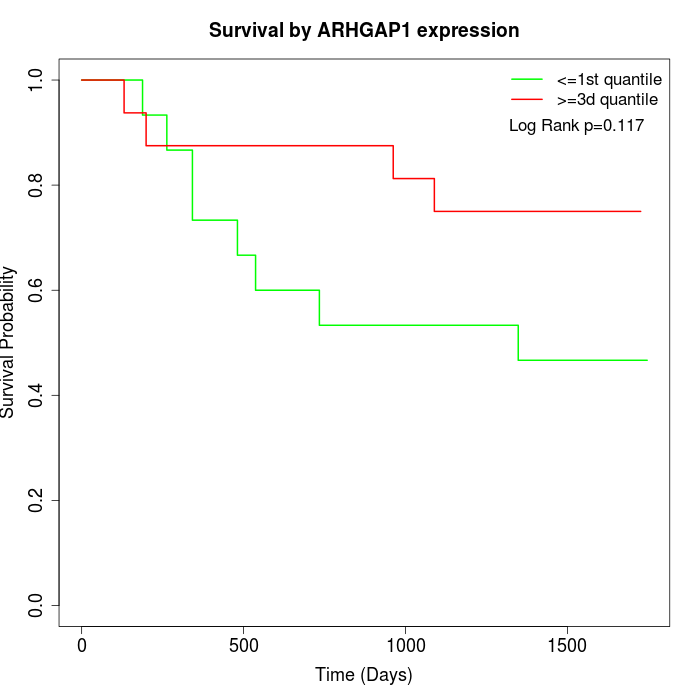
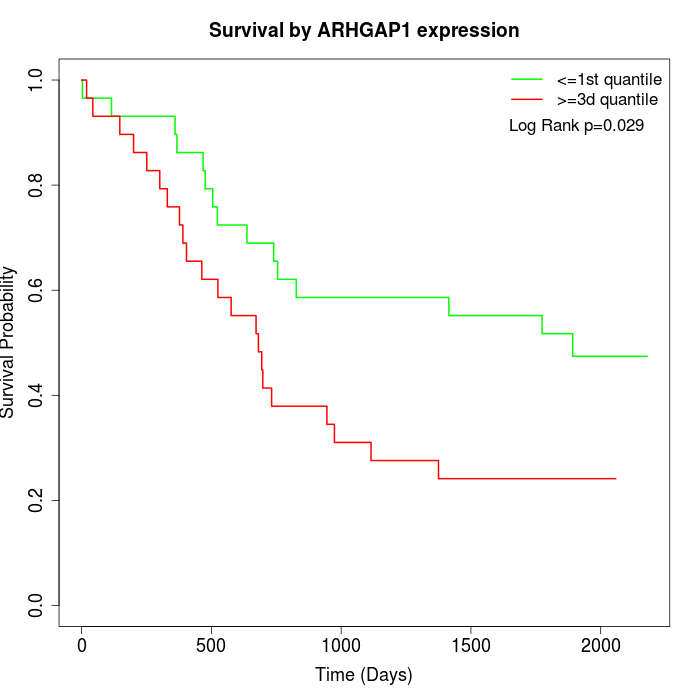
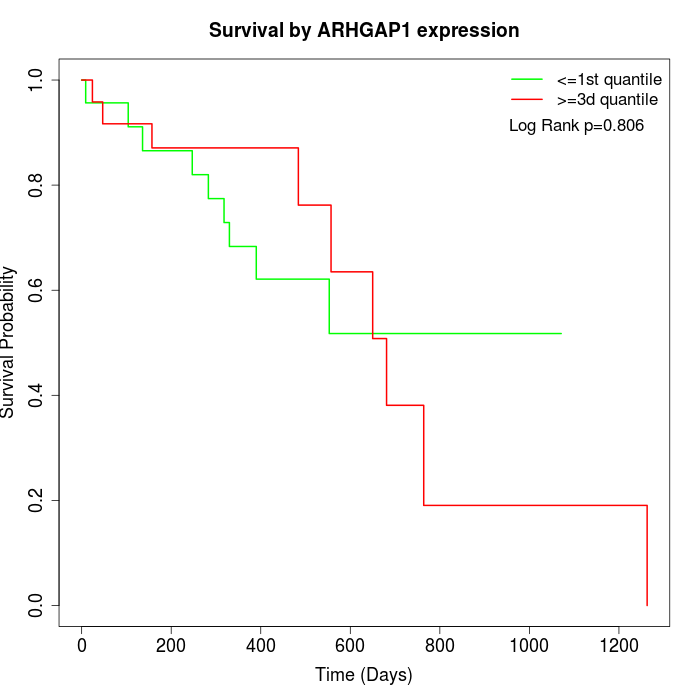
Survival by ARHGAP1 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGAP1 | 392 | 3 | 7 | 20 | |
| GSE20123 | ARHGAP1 | 392 | 3 | 6 | 21 | |
| GSE43470 | ARHGAP1 | 392 | 2 | 5 | 36 | |
| GSE46452 | ARHGAP1 | 392 | 8 | 5 | 46 | |
| GSE47630 | ARHGAP1 | 392 | 3 | 9 | 28 | |
| GSE54993 | ARHGAP1 | 392 | 3 | 0 | 67 | |
| GSE54994 | ARHGAP1 | 392 | 1 | 10 | 42 | |
| GSE60625 | ARHGAP1 | 392 | 0 | 0 | 11 | |
| GSE74703 | ARHGAP1 | 392 | 2 | 2 | 32 | |
| GSE74704 | ARHGAP1 | 392 | 2 | 4 | 14 | |
| TCGA | ARHGAP1 | 392 | 13 | 19 | 64 |
Total number of gains: 40; Total number of losses: 67; Total Number of normals: 381.
Somatic mutations of ARHGAP1:
Generating mutation plots.
Highly correlated genes for ARHGAP1:
Showing top 20/901 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP1 | AHSP | 0.7849 | 3 | 0 | 3 |
| ARHGAP1 | SCOC | 0.73548 | 3 | 0 | 3 |
| ARHGAP1 | ESYT2 | 0.733034 | 4 | 0 | 4 |
| ARHGAP1 | FRMD8 | 0.719201 | 3 | 0 | 3 |
| ARHGAP1 | ANKRD37 | 0.718996 | 3 | 0 | 3 |
| ARHGAP1 | RAB3GAP1 | 0.713653 | 3 | 0 | 3 |
| ARHGAP1 | XKR8 | 0.710853 | 3 | 0 | 3 |
| ARHGAP1 | PTGR1 | 0.703237 | 3 | 0 | 3 |
| ARHGAP1 | SH2B1 | 0.697525 | 3 | 0 | 3 |
| ARHGAP1 | KAT7 | 0.697044 | 3 | 0 | 3 |
| ARHGAP1 | CDK8 | 0.688952 | 3 | 0 | 3 |
| ARHGAP1 | PPM1L | 0.687781 | 3 | 0 | 3 |
| ARHGAP1 | KCNC4 | 0.68086 | 3 | 0 | 3 |
| ARHGAP1 | DVL1 | 0.680373 | 4 | 0 | 3 |
| ARHGAP1 | LANCL1 | 0.679232 | 4 | 0 | 4 |
| ARHGAP1 | SLC16A14 | 0.674249 | 3 | 0 | 3 |
| ARHGAP1 | CCDC186 | 0.673558 | 4 | 0 | 4 |
| ARHGAP1 | HLA-DQB2 | 0.673381 | 4 | 0 | 4 |
| ARHGAP1 | CNTF | 0.670954 | 3 | 0 | 3 |
| ARHGAP1 | GRHPR | 0.667958 | 5 | 0 | 4 |
For details and further investigation, click here