| Full name: RAB3 GTPase activating protein catalytic subunit 1 | Alias Symbol: RAB3GAP|KIAA0066|RAB3GAP130|WARBM1 | ||
| Type: protein-coding gene | Cytoband: 2q21.3 | ||
| Entrez ID: 22930 | HGNC ID: HGNC:17063 | Ensembl Gene: ENSG00000115839 | OMIM ID: 602536 |
| Drug and gene relationship at DGIdb | |||
Expression of RAB3GAP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAB3GAP1 | 22930 | 213531_s_at | -0.0212 | 0.9655 | |
| GSE20347 | RAB3GAP1 | 22930 | 213531_s_at | -0.0306 | 0.7789 | |
| GSE23400 | RAB3GAP1 | 22930 | 213531_s_at | 0.0568 | 0.2911 | |
| GSE26886 | RAB3GAP1 | 22930 | 213531_s_at | -0.0572 | 0.7959 | |
| GSE29001 | RAB3GAP1 | 22930 | 213531_s_at | 0.1383 | 0.4842 | |
| GSE38129 | RAB3GAP1 | 22930 | 213531_s_at | -0.0425 | 0.6641 | |
| GSE45670 | RAB3GAP1 | 22930 | 213531_s_at | 0.0476 | 0.6876 | |
| GSE53622 | RAB3GAP1 | 22930 | 38110 | -0.2133 | 0.0001 | |
| GSE53624 | RAB3GAP1 | 22930 | 38110 | -0.2409 | 0.0000 | |
| GSE63941 | RAB3GAP1 | 22930 | 213531_s_at | -0.2060 | 0.6279 | |
| GSE77861 | RAB3GAP1 | 22930 | 213531_s_at | 0.2073 | 0.1973 | |
| GSE97050 | RAB3GAP1 | 22930 | A_23_P50942 | -0.3201 | 0.2113 | |
| SRP007169 | RAB3GAP1 | 22930 | RNAseq | -0.2285 | 0.5165 | |
| SRP008496 | RAB3GAP1 | 22930 | RNAseq | -0.1007 | 0.7194 | |
| SRP064894 | RAB3GAP1 | 22930 | RNAseq | -0.2232 | 0.1918 | |
| SRP133303 | RAB3GAP1 | 22930 | RNAseq | -0.0104 | 0.9358 | |
| SRP159526 | RAB3GAP1 | 22930 | RNAseq | -0.1732 | 0.4770 | |
| SRP193095 | RAB3GAP1 | 22930 | RNAseq | -0.3466 | 0.0003 | |
| SRP219564 | RAB3GAP1 | 22930 | RNAseq | -0.5202 | 0.0826 | |
| TCGA | RAB3GAP1 | 22930 | RNAseq | -0.0863 | 0.0552 |
Upregulated datasets: 0; Downregulated datasets: 0.
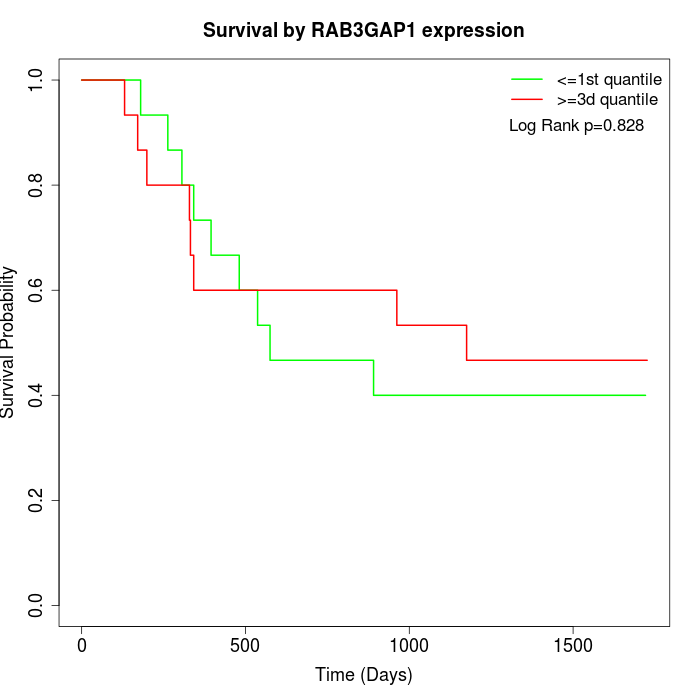
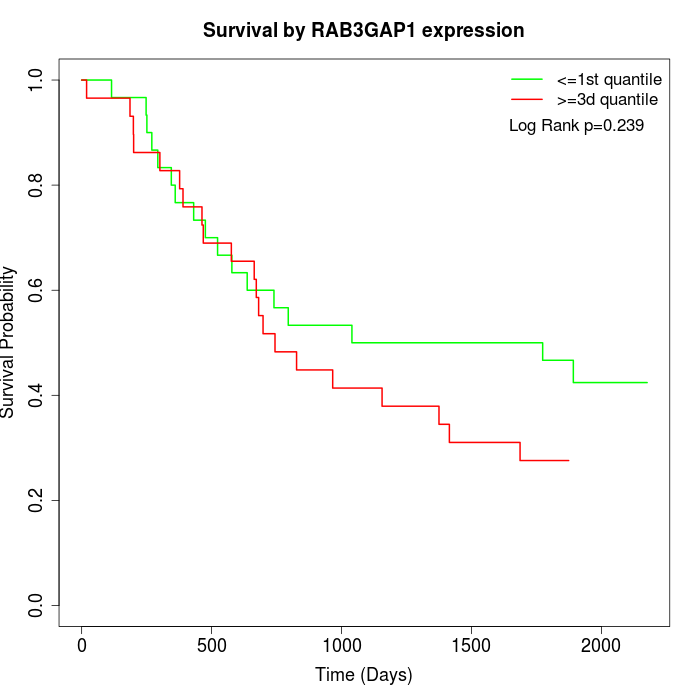
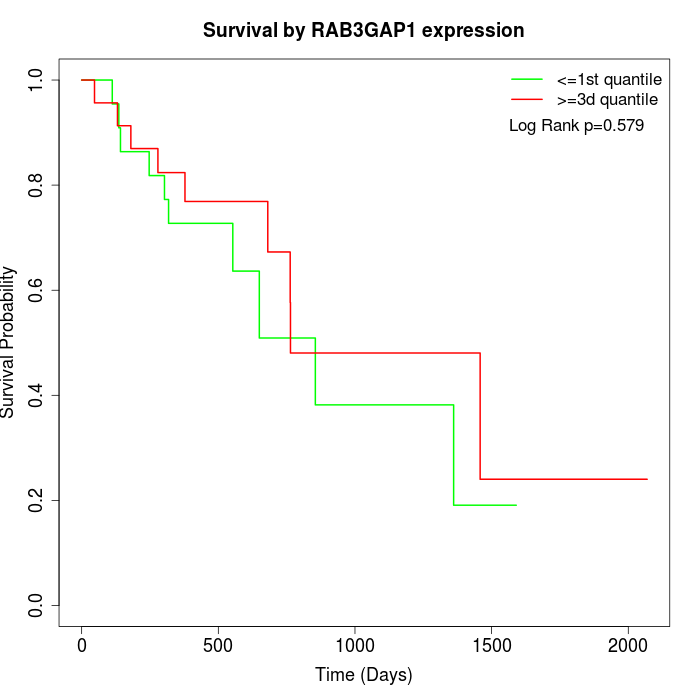
Survival by RAB3GAP1 expression:
Note: Click image to view full size file.
Copy number change of RAB3GAP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAB3GAP1 | 22930 | 2 | 4 | 24 | |
| GSE20123 | RAB3GAP1 | 22930 | 2 | 4 | 24 | |
| GSE43470 | RAB3GAP1 | 22930 | 1 | 1 | 41 | |
| GSE46452 | RAB3GAP1 | 22930 | 1 | 4 | 54 | |
| GSE47630 | RAB3GAP1 | 22930 | 5 | 2 | 33 | |
| GSE54993 | RAB3GAP1 | 22930 | 0 | 6 | 64 | |
| GSE54994 | RAB3GAP1 | 22930 | 9 | 0 | 44 | |
| GSE60625 | RAB3GAP1 | 22930 | 0 | 3 | 8 | |
| GSE74703 | RAB3GAP1 | 22930 | 1 | 1 | 34 | |
| GSE74704 | RAB3GAP1 | 22930 | 1 | 1 | 18 | |
| TCGA | RAB3GAP1 | 22930 | 21 | 10 | 65 |
Total number of gains: 43; Total number of losses: 36; Total Number of normals: 409.
Somatic mutations of RAB3GAP1:
Generating mutation plots.
Highly correlated genes for RAB3GAP1:
Showing top 20/139 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAB3GAP1 | PARP2 | 0.780231 | 3 | 0 | 3 |
| RAB3GAP1 | PPP2R3A | 0.773346 | 3 | 0 | 3 |
| RAB3GAP1 | CDADC1 | 0.759078 | 3 | 0 | 3 |
| RAB3GAP1 | SIPA1L2 | 0.751459 | 4 | 0 | 4 |
| RAB3GAP1 | TTLL11 | 0.746637 | 3 | 0 | 3 |
| RAB3GAP1 | SFPQ | 0.721789 | 3 | 0 | 3 |
| RAB3GAP1 | ZC3H15 | 0.719442 | 3 | 0 | 3 |
| RAB3GAP1 | ARHGAP1 | 0.713653 | 3 | 0 | 3 |
| RAB3GAP1 | CNOT2 | 0.71191 | 3 | 0 | 3 |
| RAB3GAP1 | KLHL20 | 0.711153 | 5 | 0 | 5 |
| RAB3GAP1 | RNF185 | 0.701104 | 3 | 0 | 3 |
| RAB3GAP1 | COMMD1 | 0.699361 | 3 | 0 | 3 |
| RAB3GAP1 | AKAP17A | 0.687412 | 3 | 0 | 3 |
| RAB3GAP1 | CCDC59 | 0.684841 | 3 | 0 | 3 |
| RAB3GAP1 | TIGD6 | 0.679346 | 3 | 0 | 3 |
| RAB3GAP1 | KCTD6 | 0.678048 | 3 | 0 | 3 |
| RAB3GAP1 | NR2C2 | 0.675557 | 3 | 0 | 3 |
| RAB3GAP1 | CCDC159 | 0.674301 | 3 | 0 | 3 |
| RAB3GAP1 | CYP26B1 | 0.672629 | 3 | 0 | 3 |
| RAB3GAP1 | ST6GALNAC6 | 0.669219 | 3 | 0 | 3 |
For details and further investigation, click here