| Full name: Rho guanine nucleotide exchange factor 26 | Alias Symbol: DKFZP434D146|SGEF | ||
| Type: protein-coding gene | Cytoband: 3q25.2 | ||
| Entrez ID: 26084 | HGNC ID: HGNC:24490 | Ensembl Gene: ENSG00000114790 | OMIM ID: 617552 |
| Drug and gene relationship at DGIdb | |||
ARHGEF26 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05100 | Bacterial invasion of epithelial cells |
Expression of ARHGEF26:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGEF26 | 26084 | 227197_at | -1.5807 | 0.2107 | |
| GSE20347 | ARHGEF26 | 26084 | 222121_at | -0.0644 | 0.4779 | |
| GSE23400 | ARHGEF26 | 26084 | 222121_at | -0.0919 | 0.0200 | |
| GSE26886 | ARHGEF26 | 26084 | 227197_at | -1.9907 | 0.0001 | |
| GSE29001 | ARHGEF26 | 26084 | 222121_at | -0.0561 | 0.5851 | |
| GSE38129 | ARHGEF26 | 26084 | 222121_at | -0.3483 | 0.0595 | |
| GSE45670 | ARHGEF26 | 26084 | 227197_at | -1.2153 | 0.0291 | |
| GSE53622 | ARHGEF26 | 26084 | 152675 | -0.9038 | 0.0000 | |
| GSE53624 | ARHGEF26 | 26084 | 152675 | -0.8725 | 0.0000 | |
| GSE63941 | ARHGEF26 | 26084 | 227197_at | 0.6172 | 0.6255 | |
| GSE77861 | ARHGEF26 | 26084 | 227197_at | -0.2866 | 0.6774 | |
| GSE97050 | ARHGEF26 | 26084 | A_23_P69100 | -0.8536 | 0.2371 | |
| SRP007169 | ARHGEF26 | 26084 | RNAseq | 0.1303 | 0.7331 | |
| SRP008496 | ARHGEF26 | 26084 | RNAseq | 0.4157 | 0.2741 | |
| SRP064894 | ARHGEF26 | 26084 | RNAseq | -1.0652 | 0.0005 | |
| SRP133303 | ARHGEF26 | 26084 | RNAseq | -0.3780 | 0.1973 | |
| SRP159526 | ARHGEF26 | 26084 | RNAseq | 0.2467 | 0.4746 | |
| SRP193095 | ARHGEF26 | 26084 | RNAseq | -0.2348 | 0.4370 | |
| SRP219564 | ARHGEF26 | 26084 | RNAseq | -1.1219 | 0.2143 |
Upregulated datasets: 0; Downregulated datasets: 3.
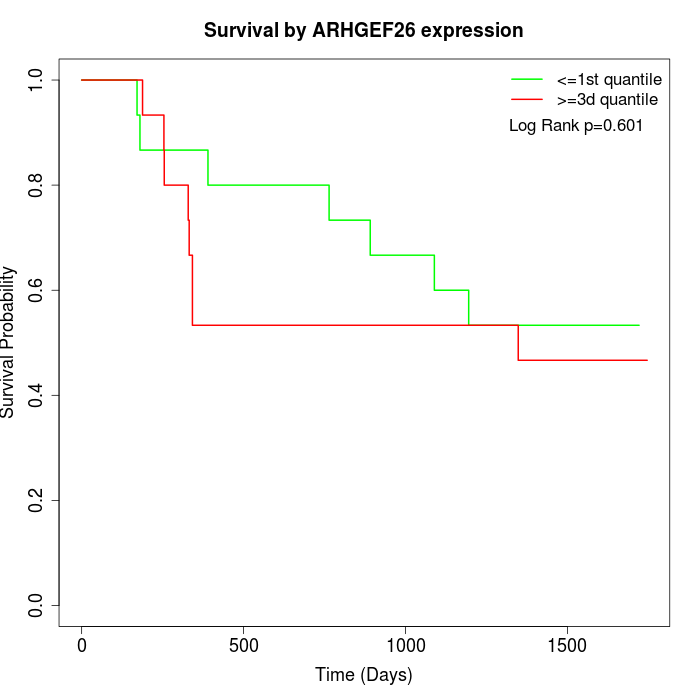
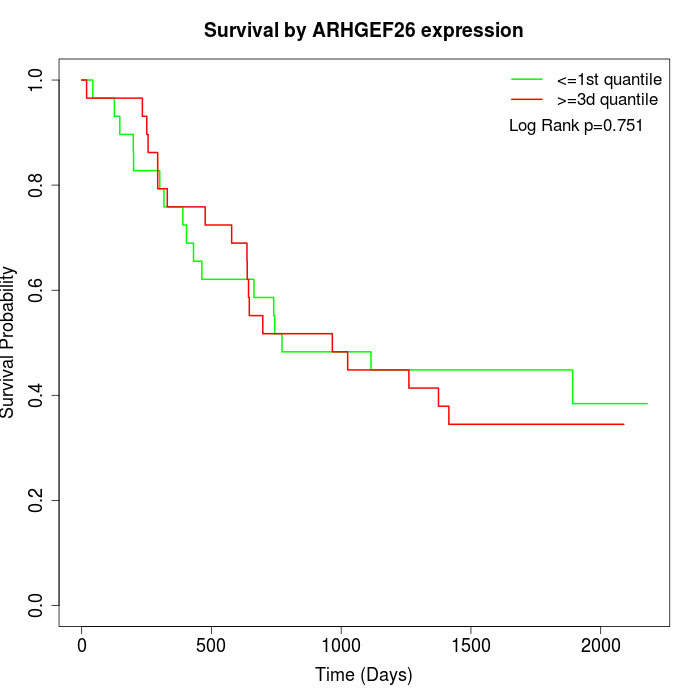
Survival by ARHGEF26 expression:
Note: Click image to view full size file.
Copy number change of ARHGEF26:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGEF26 | 26084 | 22 | 0 | 8 | |
| GSE20123 | ARHGEF26 | 26084 | 22 | 0 | 8 | |
| GSE43470 | ARHGEF26 | 26084 | 24 | 0 | 19 | |
| GSE46452 | ARHGEF26 | 26084 | 18 | 2 | 39 | |
| GSE47630 | ARHGEF26 | 26084 | 19 | 3 | 18 | |
| GSE54993 | ARHGEF26 | 26084 | 1 | 10 | 59 | |
| GSE54994 | ARHGEF26 | 26084 | 38 | 1 | 14 | |
| GSE60625 | ARHGEF26 | 26084 | 0 | 6 | 5 | |
| GSE74703 | ARHGEF26 | 26084 | 21 | 0 | 15 | |
| GSE74704 | ARHGEF26 | 26084 | 14 | 0 | 6 | |
| TCGA | ARHGEF26 | 26084 | 73 | 1 | 22 |
Total number of gains: 252; Total number of losses: 23; Total Number of normals: 213.
Somatic mutations of ARHGEF26:
Generating mutation plots.
Highly correlated genes for ARHGEF26:
Showing top 20/436 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGEF26 | GREB1L | 0.724782 | 3 | 0 | 3 |
| ARHGEF26 | ROBO2 | 0.705394 | 3 | 0 | 3 |
| ARHGEF26 | HLF | 0.70484 | 8 | 0 | 8 |
| ARHGEF26 | ID4 | 0.700968 | 4 | 0 | 3 |
| ARHGEF26 | CAMK2A | 0.689136 | 3 | 0 | 3 |
| ARHGEF26 | HDHD2 | 0.684894 | 3 | 0 | 3 |
| ARHGEF26 | B4GALT6 | 0.683134 | 4 | 0 | 4 |
| ARHGEF26 | MOB2 | 0.678961 | 5 | 0 | 5 |
| ARHGEF26 | GKAP1 | 0.676569 | 5 | 0 | 4 |
| ARHGEF26 | DAPL1 | 0.675745 | 4 | 0 | 4 |
| ARHGEF26 | FAM3B | 0.675482 | 5 | 0 | 5 |
| ARHGEF26 | FAM13C | 0.673643 | 4 | 0 | 4 |
| ARHGEF26 | SLC9A9 | 0.671417 | 6 | 0 | 5 |
| ARHGEF26 | LONRF2 | 0.670962 | 4 | 0 | 3 |
| ARHGEF26 | ZNF471 | 0.665651 | 5 | 0 | 5 |
| ARHGEF26 | CLDN3 | 0.661513 | 3 | 0 | 3 |
| ARHGEF26 | PLCB4 | 0.660908 | 7 | 0 | 7 |
| ARHGEF26 | TACC1 | 0.658692 | 9 | 0 | 8 |
| ARHGEF26 | FAM219B | 0.654783 | 3 | 0 | 3 |
| ARHGEF26 | ICA1L | 0.650101 | 4 | 0 | 4 |
For details and further investigation, click here