| Full name: ankyrin repeat and SOCS box containing 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 136371 | HGNC ID: HGNC:17185 | Ensembl Gene: ENSG00000146926 | OMIM ID: 615054 |
| Drug and gene relationship at DGIdb | |||
Expression of ASB10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ASB10 | 136371 | 1553039_a_at | -0.0352 | 0.8948 | |
| GSE26886 | ASB10 | 136371 | 1553039_a_at | 0.0285 | 0.8600 | |
| GSE45670 | ASB10 | 136371 | 1553039_a_at | -0.0295 | 0.7468 | |
| GSE53622 | ASB10 | 136371 | 42716 | 0.0834 | 0.1774 | |
| GSE53624 | ASB10 | 136371 | 42716 | 0.2496 | 0.0001 | |
| GSE63941 | ASB10 | 136371 | 1553039_a_at | 0.1975 | 0.2194 | |
| GSE77861 | ASB10 | 136371 | 1553039_a_at | -0.2315 | 0.1999 | |
| GSE97050 | ASB10 | 136371 | A_33_P3290124 | -0.0051 | 0.9866 |
Upregulated datasets: 0; Downregulated datasets: 0.
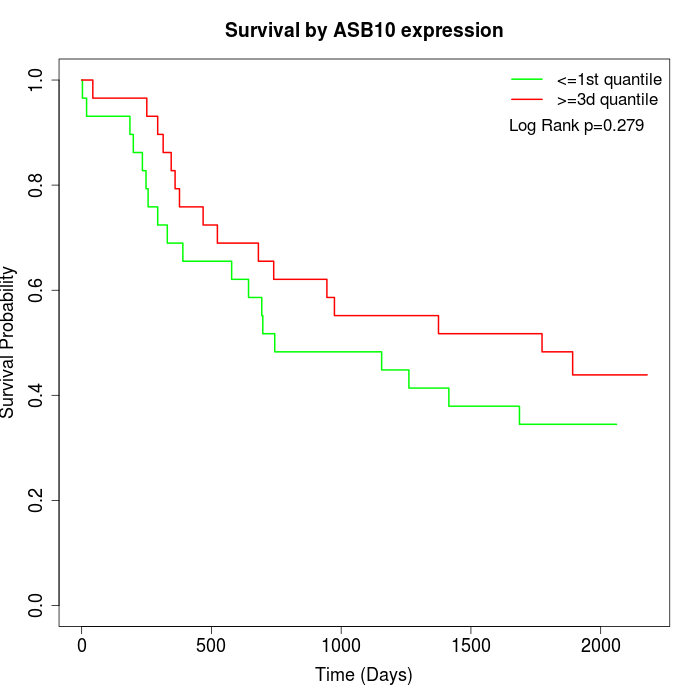
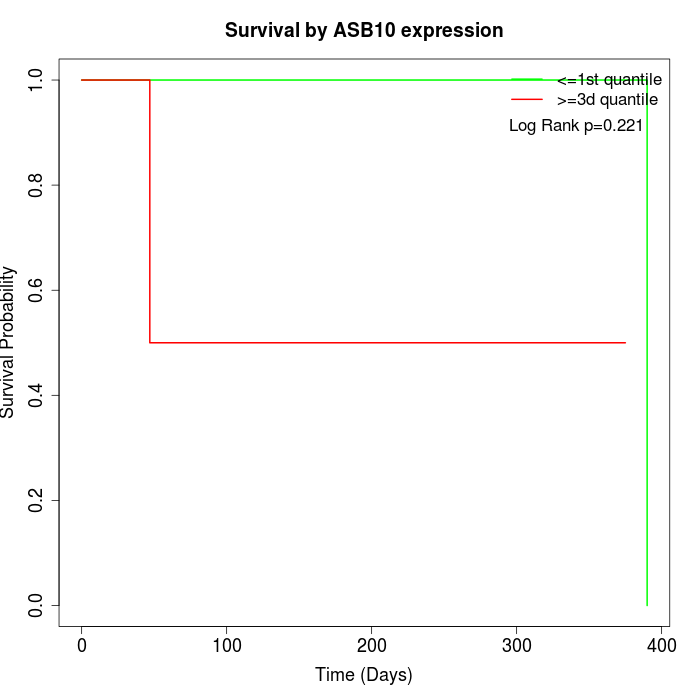
Survival by ASB10 expression:
Note: Click image to view full size file.
Copy number change of ASB10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ASB10 | 136371 | 2 | 4 | 24 | |
| GSE20123 | ASB10 | 136371 | 2 | 4 | 24 | |
| GSE43470 | ASB10 | 136371 | 2 | 5 | 36 | |
| GSE46452 | ASB10 | 136371 | 7 | 2 | 50 | |
| GSE47630 | ASB10 | 136371 | 5 | 9 | 26 | |
| GSE54993 | ASB10 | 136371 | 3 | 3 | 64 | |
| GSE54994 | ASB10 | 136371 | 5 | 8 | 40 | |
| GSE60625 | ASB10 | 136371 | 0 | 0 | 11 | |
| GSE74703 | ASB10 | 136371 | 2 | 4 | 30 | |
| GSE74704 | ASB10 | 136371 | 1 | 4 | 15 | |
| TCGA | ASB10 | 136371 | 28 | 25 | 43 |
Total number of gains: 57; Total number of losses: 68; Total Number of normals: 363.
Somatic mutations of ASB10:
Generating mutation plots.
Highly correlated genes for ASB10:
Showing top 20/426 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ASB10 | NRSN1 | 0.756369 | 3 | 0 | 3 |
| ASB10 | ADCY1 | 0.751919 | 3 | 0 | 3 |
| ASB10 | TMEM59L | 0.743014 | 3 | 0 | 3 |
| ASB10 | SPACA3 | 0.741487 | 3 | 0 | 3 |
| ASB10 | ATP2B3 | 0.720181 | 3 | 0 | 3 |
| ASB10 | CALCA | 0.716467 | 3 | 0 | 3 |
| ASB10 | PABPN1L | 0.712492 | 3 | 0 | 3 |
| ASB10 | VIPR2 | 0.711255 | 4 | 0 | 3 |
| ASB10 | ESAM | 0.710142 | 3 | 0 | 3 |
| ASB10 | GRIN2C | 0.70728 | 3 | 0 | 3 |
| ASB10 | ITGA10 | 0.699826 | 3 | 0 | 3 |
| ASB10 | TCTE3 | 0.699772 | 3 | 0 | 3 |
| ASB10 | CCL17 | 0.698474 | 3 | 0 | 3 |
| ASB10 | C22orf23 | 0.693086 | 4 | 0 | 3 |
| ASB10 | USH1C | 0.690707 | 5 | 0 | 5 |
| ASB10 | DPP6 | 0.689458 | 4 | 0 | 4 |
| ASB10 | NXNL1 | 0.685165 | 4 | 0 | 3 |
| ASB10 | IZUMO2 | 0.68346 | 3 | 0 | 3 |
| ASB10 | ARL4D | 0.683279 | 3 | 0 | 3 |
| ASB10 | STX1B | 0.68155 | 4 | 0 | 3 |
For details and further investigation, click here