| Full name: BCL2 related protein A1 | Alias Symbol: GRS|BFL1|BCL2L5|ACC-1|ACC-2|ACC2|ACC1 | ||
| Type: protein-coding gene | Cytoband: 15q25.1 | ||
| Entrez ID: 597 | HGNC ID: HGNC:991 | Ensembl Gene: ENSG00000140379 | OMIM ID: 601056 |
| Drug and gene relationship at DGIdb | |||
BCL2A1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04210 | Apoptosis |
Expression of BCL2A1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCL2A1 | 597 | 205681_at | 1.7128 | 0.2507 | |
| GSE20347 | BCL2A1 | 597 | 205681_at | 0.9210 | 0.0741 | |
| GSE23400 | BCL2A1 | 597 | 205681_at | 0.4324 | 0.0002 | |
| GSE26886 | BCL2A1 | 597 | 205681_at | 0.8768 | 0.0208 | |
| GSE29001 | BCL2A1 | 597 | 205681_at | 0.4394 | 0.0322 | |
| GSE38129 | BCL2A1 | 597 | 205681_at | 1.0775 | 0.0099 | |
| GSE45670 | BCL2A1 | 597 | 205681_at | 0.8937 | 0.0867 | |
| GSE53622 | BCL2A1 | 597 | 156671 | 1.9197 | 0.0000 | |
| GSE53624 | BCL2A1 | 597 | 156671 | 1.4184 | 0.0000 | |
| GSE63941 | BCL2A1 | 597 | 205681_at | -0.9508 | 0.3042 | |
| GSE77861 | BCL2A1 | 597 | 205681_at | 0.5526 | 0.0084 | |
| GSE97050 | BCL2A1 | 597 | A_23_P321703 | 0.6687 | 0.1972 | |
| SRP007169 | BCL2A1 | 597 | RNAseq | 2.7967 | 0.0017 | |
| SRP008496 | BCL2A1 | 597 | RNAseq | 2.1744 | 0.0004 | |
| SRP064894 | BCL2A1 | 597 | RNAseq | 1.9268 | 0.0000 | |
| SRP133303 | BCL2A1 | 597 | RNAseq | 2.5709 | 0.0000 | |
| SRP159526 | BCL2A1 | 597 | RNAseq | 2.9855 | 0.0031 | |
| SRP193095 | BCL2A1 | 597 | RNAseq | 2.3290 | 0.0000 | |
| SRP219564 | BCL2A1 | 597 | RNAseq | 1.6352 | 0.0061 | |
| TCGA | BCL2A1 | 597 | RNAseq | 1.1040 | 0.0000 |
Upregulated datasets: 11; Downregulated datasets: 0.
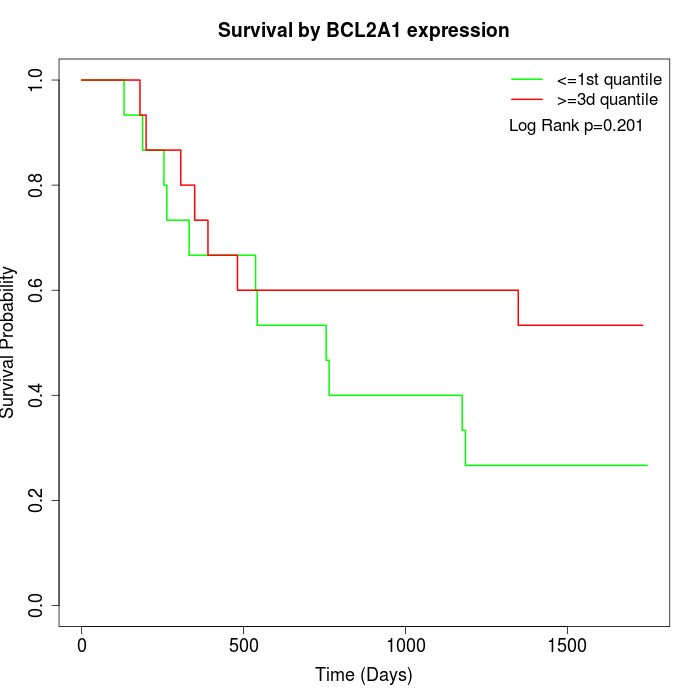
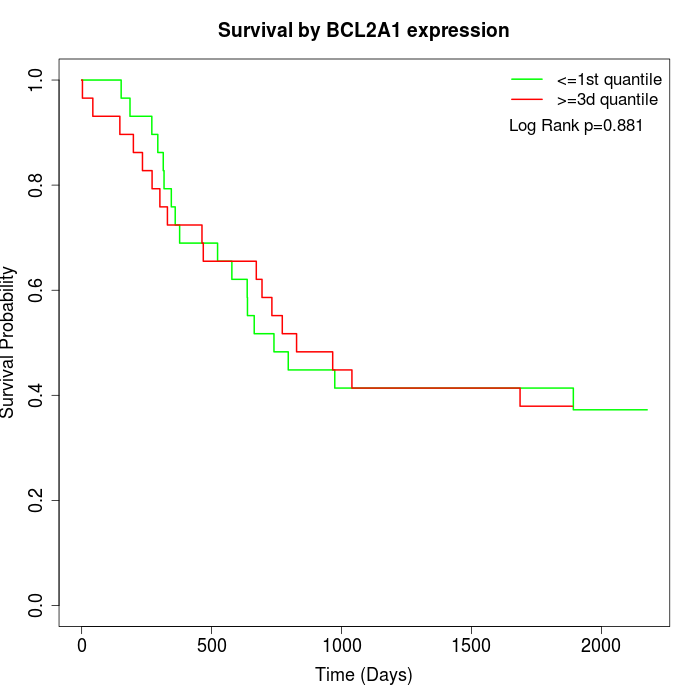
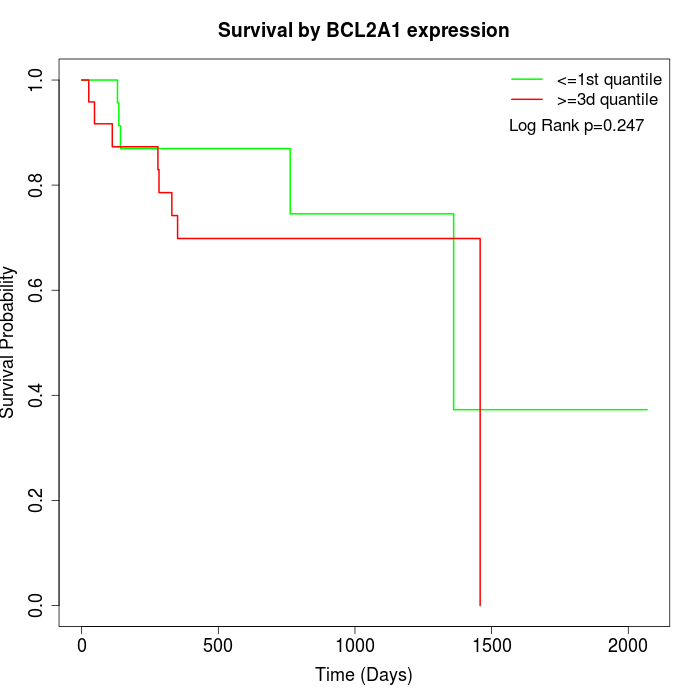
Survival by BCL2A1 expression:
Note: Click image to view full size file.
Copy number change of BCL2A1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCL2A1 | 597 | 7 | 1 | 22 | |
| GSE20123 | BCL2A1 | 597 | 7 | 1 | 22 | |
| GSE43470 | BCL2A1 | 597 | 4 | 4 | 35 | |
| GSE46452 | BCL2A1 | 597 | 3 | 7 | 49 | |
| GSE47630 | BCL2A1 | 597 | 8 | 10 | 22 | |
| GSE54993 | BCL2A1 | 597 | 5 | 6 | 59 | |
| GSE54994 | BCL2A1 | 597 | 5 | 6 | 42 | |
| GSE60625 | BCL2A1 | 597 | 4 | 0 | 7 | |
| GSE74703 | BCL2A1 | 597 | 4 | 2 | 30 | |
| GSE74704 | BCL2A1 | 597 | 3 | 1 | 16 | |
| TCGA | BCL2A1 | 597 | 15 | 15 | 66 |
Total number of gains: 65; Total number of losses: 53; Total Number of normals: 370.
Somatic mutations of BCL2A1:
Generating mutation plots.
Highly correlated genes for BCL2A1:
Showing top 20/1294 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCL2A1 | FCGR3A | 0.837625 | 3 | 0 | 3 |
| BCL2A1 | HELB | 0.780975 | 3 | 0 | 3 |
| BCL2A1 | MEX3B | 0.751864 | 4 | 0 | 4 |
| BCL2A1 | LRRC8C | 0.747008 | 3 | 0 | 3 |
| BCL2A1 | PROK2 | 0.746522 | 6 | 0 | 6 |
| BCL2A1 | CCDC127 | 0.738509 | 3 | 0 | 3 |
| BCL2A1 | CLEC4E | 0.736755 | 3 | 0 | 3 |
| BCL2A1 | MGAT5 | 0.735267 | 4 | 0 | 3 |
| BCL2A1 | SUCNR1 | 0.730198 | 4 | 0 | 4 |
| BCL2A1 | INTS10 | 0.728705 | 3 | 0 | 3 |
| BCL2A1 | C19orf38 | 0.721115 | 3 | 0 | 3 |
| BCL2A1 | ZNF827 | 0.719837 | 4 | 0 | 4 |
| BCL2A1 | CERK | 0.719416 | 4 | 0 | 3 |
| BCL2A1 | FAM167B | 0.716313 | 3 | 0 | 3 |
| BCL2A1 | RAB34 | 0.714255 | 3 | 0 | 3 |
| BCL2A1 | TMEM60 | 0.710984 | 3 | 0 | 3 |
| BCL2A1 | CD163L1 | 0.710692 | 3 | 0 | 3 |
| BCL2A1 | SLC36A1 | 0.709481 | 3 | 0 | 3 |
| BCL2A1 | KHSRP | 0.709413 | 4 | 0 | 3 |
| BCL2A1 | CYP20A1 | 0.707602 | 3 | 0 | 3 |
For details and further investigation, click here