| Full name: succinate receptor 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q25.1 | ||
| Entrez ID: 56670 | HGNC ID: HGNC:4542 | Ensembl Gene: ENSG00000198829 | OMIM ID: 606381 |
| Drug and gene relationship at DGIdb | |||
SUCNR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway |
Expression of SUCNR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SUCNR1 | 56670 | 223939_at | 0.2802 | 0.5431 | |
| GSE26886 | SUCNR1 | 56670 | 223939_at | -0.1490 | 0.2039 | |
| GSE45670 | SUCNR1 | 56670 | 223939_at | 0.0324 | 0.8525 | |
| GSE53622 | SUCNR1 | 56670 | 14841 | 1.6317 | 0.0000 | |
| GSE53624 | SUCNR1 | 56670 | 14841 | 1.6326 | 0.0000 | |
| GSE63941 | SUCNR1 | 56670 | 223939_at | -0.1301 | 0.5879 | |
| GSE77861 | SUCNR1 | 56670 | 223939_at | -0.0180 | 0.8896 | |
| GSE97050 | SUCNR1 | 56670 | A_24_P265832 | 0.7217 | 0.1159 | |
| SRP007169 | SUCNR1 | 56670 | RNAseq | 2.6707 | 0.0058 | |
| SRP133303 | SUCNR1 | 56670 | RNAseq | 1.9310 | 0.0000 | |
| SRP159526 | SUCNR1 | 56670 | RNAseq | 1.9074 | 0.0259 | |
| TCGA | SUCNR1 | 56670 | RNAseq | 1.4457 | 0.0016 |
Upregulated datasets: 6; Downregulated datasets: 0.
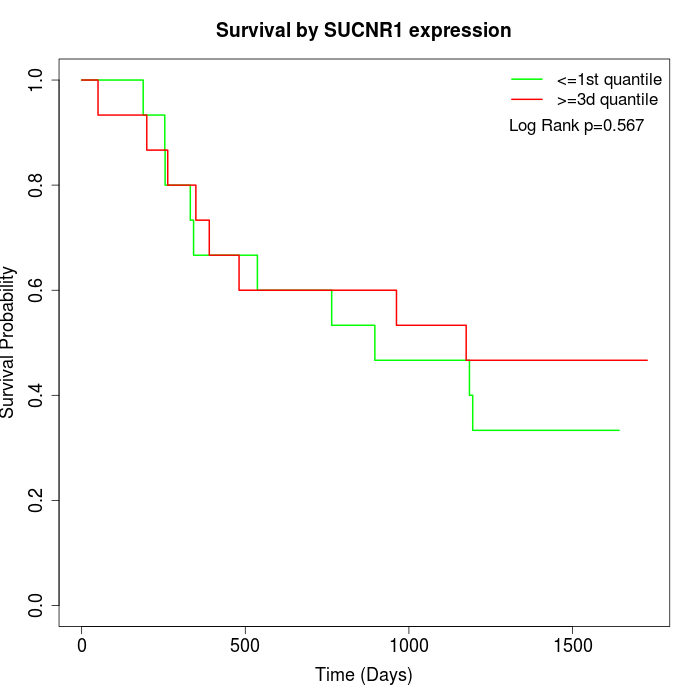
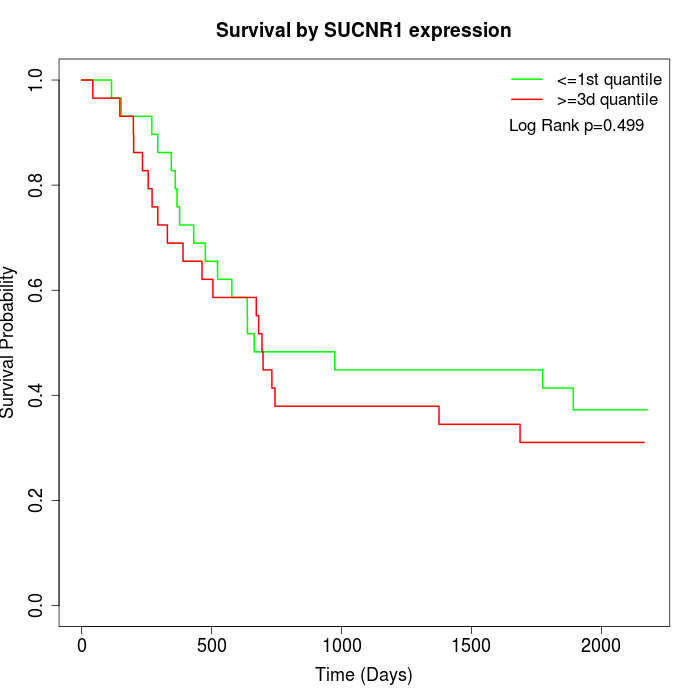
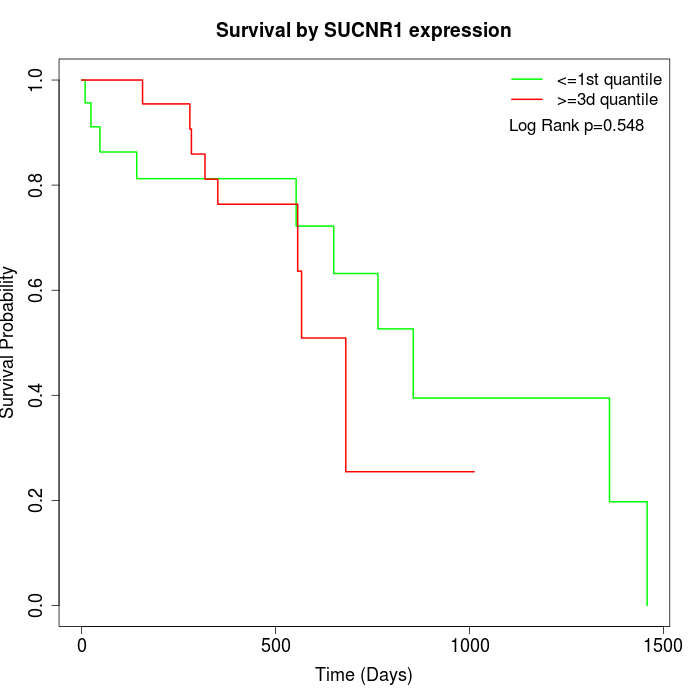
Survival by SUCNR1 expression:
Note: Click image to view full size file.
Copy number change of SUCNR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SUCNR1 | 56670 | 21 | 0 | 9 | |
| GSE20123 | SUCNR1 | 56670 | 21 | 0 | 9 | |
| GSE43470 | SUCNR1 | 56670 | 23 | 0 | 20 | |
| GSE46452 | SUCNR1 | 56670 | 18 | 2 | 39 | |
| GSE47630 | SUCNR1 | 56670 | 19 | 3 | 18 | |
| GSE54993 | SUCNR1 | 56670 | 1 | 10 | 59 | |
| GSE54994 | SUCNR1 | 56670 | 37 | 1 | 15 | |
| GSE60625 | SUCNR1 | 56670 | 0 | 6 | 5 | |
| GSE74703 | SUCNR1 | 56670 | 20 | 0 | 16 | |
| GSE74704 | SUCNR1 | 56670 | 14 | 0 | 6 | |
| TCGA | SUCNR1 | 56670 | 73 | 1 | 22 |
Total number of gains: 247; Total number of losses: 23; Total Number of normals: 218.
Somatic mutations of SUCNR1:
Generating mutation plots.
Highly correlated genes for SUCNR1:
Showing top 20/471 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SUCNR1 | LCP2 | 0.796226 | 4 | 0 | 4 |
| SUCNR1 | TREM2 | 0.783807 | 3 | 0 | 3 |
| SUCNR1 | FCGR3A | 0.782429 | 3 | 0 | 3 |
| SUCNR1 | PIK3AP1 | 0.782244 | 4 | 0 | 4 |
| SUCNR1 | TIMP1 | 0.765587 | 4 | 0 | 4 |
| SUCNR1 | ITGAM | 0.756616 | 4 | 0 | 4 |
| SUCNR1 | LRRC8C | 0.75369 | 3 | 0 | 3 |
| SUCNR1 | CMPK2 | 0.753473 | 3 | 0 | 3 |
| SUCNR1 | GIMAP6 | 0.746232 | 3 | 0 | 3 |
| SUCNR1 | GLRX | 0.7431 | 4 | 0 | 4 |
| SUCNR1 | HHEX | 0.738347 | 3 | 0 | 3 |
| SUCNR1 | BCL2A1 | 0.730198 | 4 | 0 | 4 |
| SUCNR1 | CDH11 | 0.728181 | 4 | 0 | 4 |
| SUCNR1 | TMEM176A | 0.725805 | 4 | 0 | 4 |
| SUCNR1 | SLC15A3 | 0.724962 | 4 | 0 | 4 |
| SUCNR1 | C5AR1 | 0.723992 | 4 | 0 | 4 |
| SUCNR1 | TNFRSF4 | 0.72363 | 4 | 0 | 4 |
| SUCNR1 | TLR2 | 0.722332 | 4 | 0 | 4 |
| SUCNR1 | STAT1 | 0.720562 | 3 | 0 | 3 |
| SUCNR1 | DERL3 | 0.71672 | 3 | 0 | 3 |
For details and further investigation, click here