| Full name: BCL2 like 1 | Alias Symbol: BCLX|BCL2L|Bcl-X|bcl-xL|bcl-xS|PPP1R52 | ||
| Type: protein-coding gene | Cytoband: 20q11.21 | ||
| Entrez ID: 598 | HGNC ID: HGNC:992 | Ensembl Gene: ENSG00000171552 | OMIM ID: 600039 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BCL2L1 involved pathways:
Expression of BCL2L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCL2L1 | 598 | 212312_at | 0.2855 | 0.4665 | |
| GSE20347 | BCL2L1 | 598 | 212312_at | -0.1944 | 0.1173 | |
| GSE23400 | BCL2L1 | 598 | 212312_at | -0.2118 | 0.0003 | |
| GSE26886 | BCL2L1 | 598 | 212312_at | -0.9268 | 0.0000 | |
| GSE29001 | BCL2L1 | 598 | 212312_at | -0.1097 | 0.7262 | |
| GSE38129 | BCL2L1 | 598 | 212312_at | -0.0466 | 0.6574 | |
| GSE45670 | BCL2L1 | 598 | 212312_at | 0.0170 | 0.9181 | |
| GSE53622 | BCL2L1 | 598 | 99367 | -0.3388 | 0.0003 | |
| GSE53624 | BCL2L1 | 598 | 23001 | -0.3819 | 0.0000 | |
| GSE63941 | BCL2L1 | 598 | 212312_at | 0.4160 | 0.2481 | |
| GSE77861 | BCL2L1 | 598 | 212312_at | 0.0723 | 0.7252 | |
| GSE97050 | BCL2L1 | 598 | A_23_P210886 | 0.4912 | 0.2429 | |
| SRP007169 | BCL2L1 | 598 | RNAseq | -1.2978 | 0.0140 | |
| SRP008496 | BCL2L1 | 598 | RNAseq | -1.1339 | 0.0004 | |
| SRP064894 | BCL2L1 | 598 | RNAseq | -0.7727 | 0.0000 | |
| SRP133303 | BCL2L1 | 598 | RNAseq | -0.2618 | 0.0482 | |
| SRP159526 | BCL2L1 | 598 | RNAseq | -0.8006 | 0.0017 | |
| SRP193095 | BCL2L1 | 598 | RNAseq | -0.6572 | 0.0003 | |
| SRP219564 | BCL2L1 | 598 | RNAseq | -0.6561 | 0.0307 | |
| TCGA | BCL2L1 | 598 | RNAseq | -0.1033 | 0.0461 |
Upregulated datasets: 0; Downregulated datasets: 2.
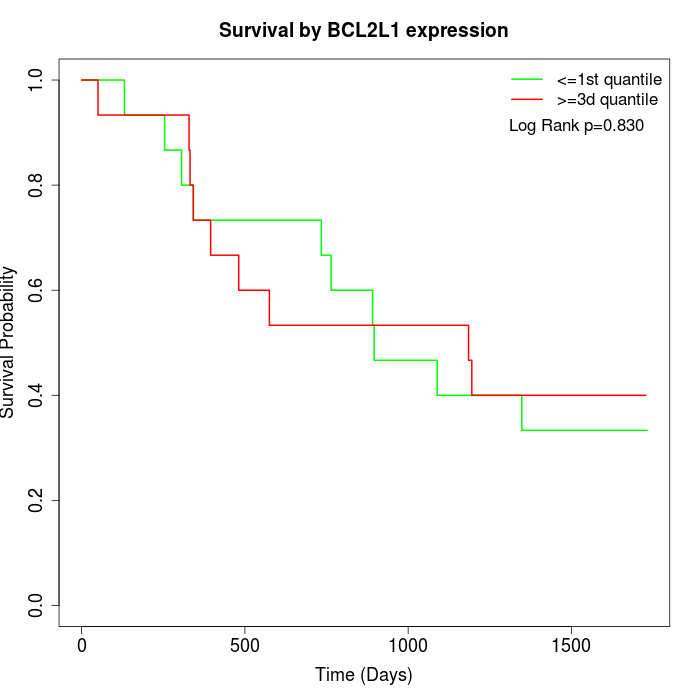
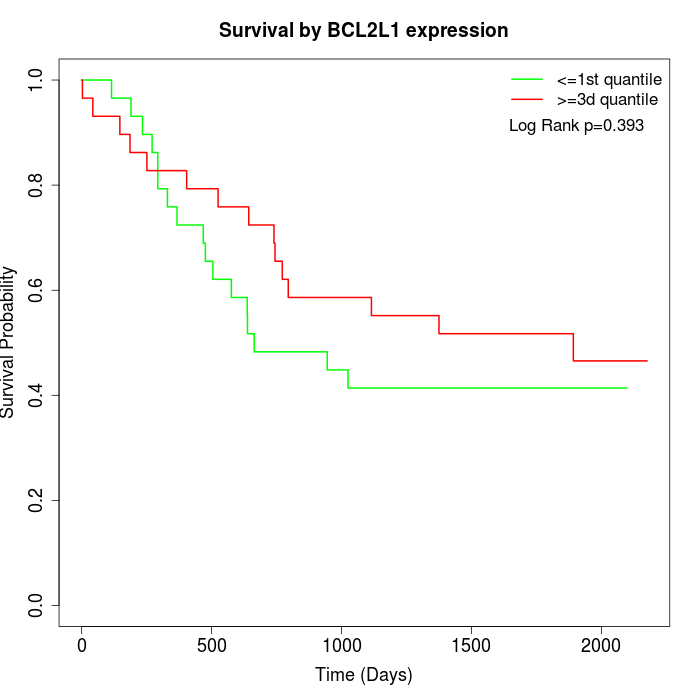
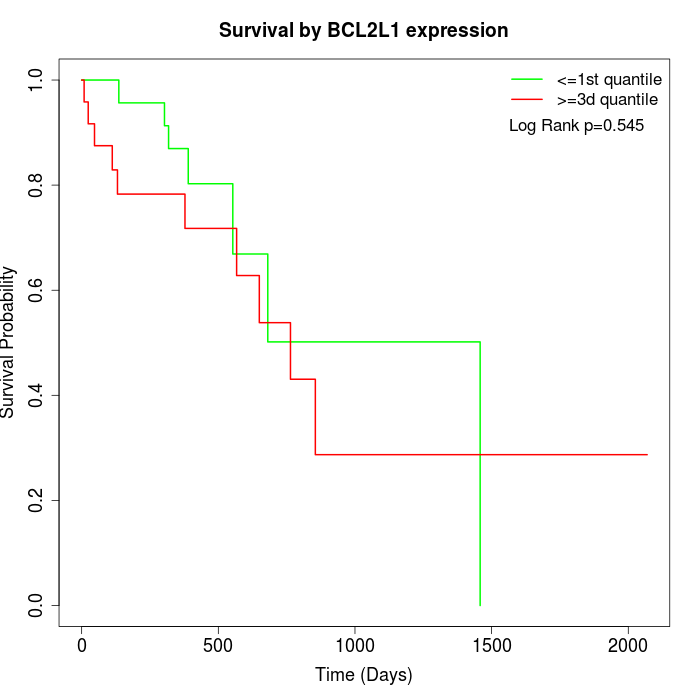
Survival by BCL2L1 expression:
Note: Click image to view full size file.
Copy number change of BCL2L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCL2L1 | 598 | 14 | 0 | 16 | |
| GSE20123 | BCL2L1 | 598 | 14 | 0 | 16 | |
| GSE43470 | BCL2L1 | 598 | 11 | 0 | 32 | |
| GSE46452 | BCL2L1 | 598 | 29 | 0 | 30 | |
| GSE47630 | BCL2L1 | 598 | 25 | 0 | 15 | |
| GSE54993 | BCL2L1 | 598 | 0 | 17 | 53 | |
| GSE54994 | BCL2L1 | 598 | 27 | 1 | 25 | |
| GSE60625 | BCL2L1 | 598 | 0 | 0 | 11 | |
| GSE74703 | BCL2L1 | 598 | 10 | 0 | 26 | |
| GSE74704 | BCL2L1 | 598 | 9 | 0 | 11 | |
| TCGA | BCL2L1 | 598 | 49 | 1 | 46 |
Total number of gains: 188; Total number of losses: 19; Total Number of normals: 281.
Somatic mutations of BCL2L1:
Generating mutation plots.
Highly correlated genes for BCL2L1:
Showing top 20/506 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCL2L1 | TMEM86B | 0.781516 | 3 | 0 | 3 |
| BCL2L1 | YTHDF2 | 0.748577 | 3 | 0 | 3 |
| BCL2L1 | PHF23 | 0.729091 | 3 | 0 | 3 |
| BCL2L1 | TRUB2 | 0.724774 | 3 | 0 | 3 |
| BCL2L1 | UBE2J2 | 0.715121 | 4 | 0 | 3 |
| BCL2L1 | INO80B | 0.713362 | 3 | 0 | 3 |
| BCL2L1 | KCNK6 | 0.70997 | 4 | 0 | 3 |
| BCL2L1 | PROSER1 | 0.705988 | 3 | 0 | 3 |
| BCL2L1 | BAIAP2L1 | 0.703329 | 3 | 0 | 3 |
| BCL2L1 | COMTD1 | 0.70141 | 3 | 0 | 3 |
| BCL2L1 | PUSL1 | 0.699322 | 4 | 0 | 4 |
| BCL2L1 | RNF187 | 0.697741 | 3 | 0 | 3 |
| BCL2L1 | LRP11 | 0.694539 | 4 | 0 | 4 |
| BCL2L1 | ANKZF1 | 0.692164 | 3 | 0 | 3 |
| BCL2L1 | GPR160 | 0.686818 | 3 | 0 | 3 |
| BCL2L1 | RAB2A | 0.678747 | 3 | 0 | 3 |
| BCL2L1 | NARF | 0.674653 | 3 | 0 | 3 |
| BCL2L1 | KCNJ5 | 0.672756 | 3 | 0 | 3 |
| BCL2L1 | TSPAN17 | 0.672284 | 3 | 0 | 3 |
| BCL2L1 | PACS1 | 0.672202 | 3 | 0 | 3 |
For details and further investigation, click here