| Full name: potassium inwardly rectifying channel subfamily J member 5 | Alias Symbol: Kir3.4|CIR|KATP1|GIRK4|LQT13 | ||
| Type: protein-coding gene | Cytoband: 11q24.3 | ||
| Entrez ID: 3762 | HGNC ID: HGNC:6266 | Ensembl Gene: ENSG00000120457 | OMIM ID: 600734 |
| Drug and gene relationship at DGIdb | |||
KCNJ5 involved pathways:
Expression of KCNJ5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ5 | 3762 | 208404_x_at | -0.0881 | 0.6789 | |
| GSE20347 | KCNJ5 | 3762 | 208404_x_at | -0.0311 | 0.6999 | |
| GSE23400 | KCNJ5 | 3762 | 211304_x_at | -0.1825 | 0.0000 | |
| GSE26886 | KCNJ5 | 3762 | 211304_x_at | -0.5682 | 0.0002 | |
| GSE29001 | KCNJ5 | 3762 | 211304_x_at | -0.1439 | 0.4259 | |
| GSE38129 | KCNJ5 | 3762 | 208404_x_at | -0.1643 | 0.1282 | |
| GSE45670 | KCNJ5 | 3762 | 211304_x_at | -0.0626 | 0.6291 | |
| GSE53622 | KCNJ5 | 3762 | 71423 | -0.3396 | 0.0691 | |
| GSE53624 | KCNJ5 | 3762 | 71423 | -0.2655 | 0.0980 | |
| GSE63941 | KCNJ5 | 3762 | 208404_x_at | 0.1199 | 0.3625 | |
| GSE77861 | KCNJ5 | 3762 | 208397_x_at | -0.2197 | 0.0539 | |
| GSE97050 | KCNJ5 | 3762 | A_24_P309521 | -0.0160 | 0.9743 | |
| SRP007169 | KCNJ5 | 3762 | RNAseq | 1.1224 | 0.1440 | |
| SRP064894 | KCNJ5 | 3762 | RNAseq | -0.0339 | 0.9157 | |
| SRP133303 | KCNJ5 | 3762 | RNAseq | -0.5910 | 0.0002 | |
| SRP159526 | KCNJ5 | 3762 | RNAseq | -1.1428 | 0.0206 | |
| SRP193095 | KCNJ5 | 3762 | RNAseq | -0.6757 | 0.0001 | |
| SRP219564 | KCNJ5 | 3762 | RNAseq | -0.7932 | 0.0428 | |
| TCGA | KCNJ5 | 3762 | RNAseq | -0.3050 | 0.4045 |
Upregulated datasets: 0; Downregulated datasets: 1.
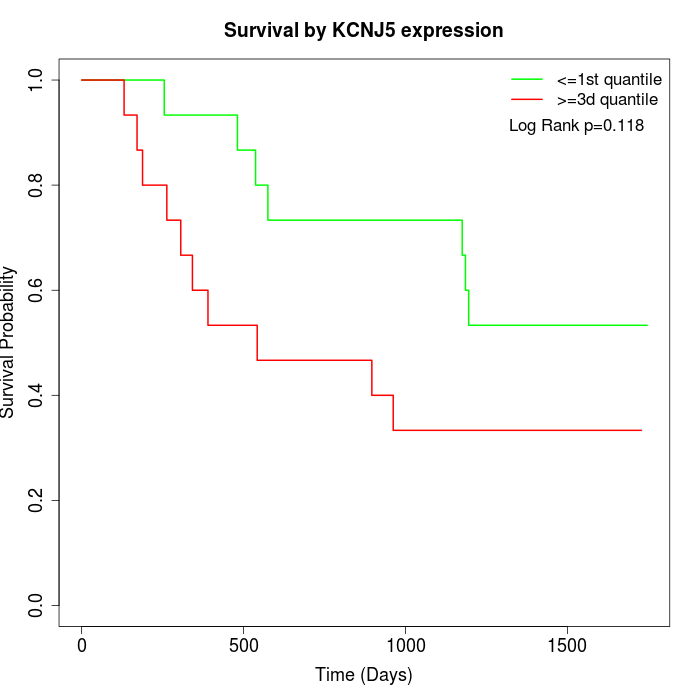
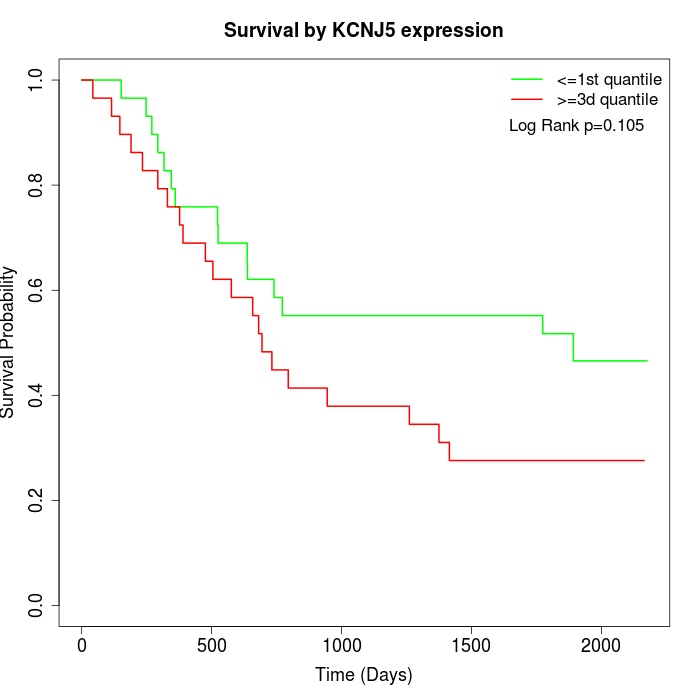
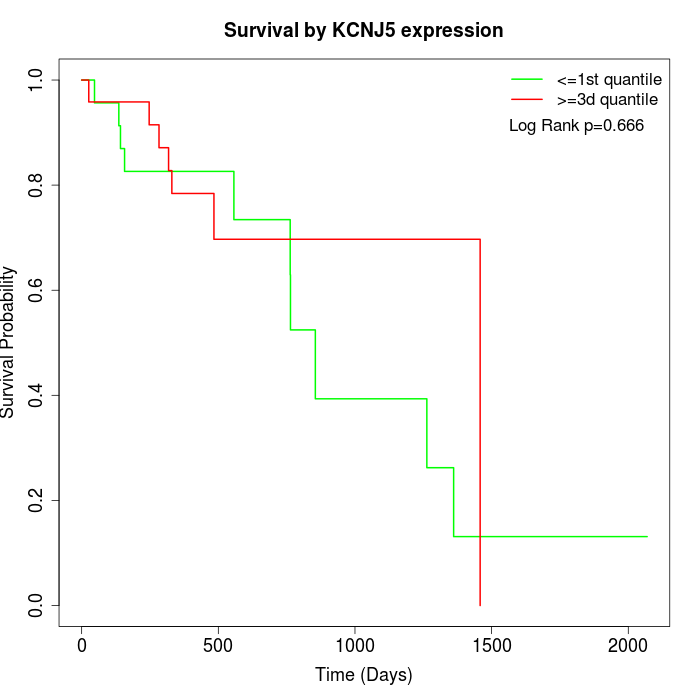
Survival by KCNJ5 expression:
Note: Click image to view full size file.
Copy number change of KCNJ5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ5 | 3762 | 0 | 13 | 17 | |
| GSE20123 | KCNJ5 | 3762 | 0 | 13 | 17 | |
| GSE43470 | KCNJ5 | 3762 | 2 | 7 | 34 | |
| GSE46452 | KCNJ5 | 3762 | 3 | 26 | 30 | |
| GSE47630 | KCNJ5 | 3762 | 2 | 19 | 19 | |
| GSE54993 | KCNJ5 | 3762 | 10 | 0 | 60 | |
| GSE54994 | KCNJ5 | 3762 | 5 | 19 | 29 | |
| GSE60625 | KCNJ5 | 3762 | 0 | 3 | 8 | |
| GSE74703 | KCNJ5 | 3762 | 1 | 5 | 30 | |
| GSE74704 | KCNJ5 | 3762 | 0 | 9 | 11 | |
| TCGA | KCNJ5 | 3762 | 6 | 50 | 40 |
Total number of gains: 29; Total number of losses: 164; Total Number of normals: 295.
Somatic mutations of KCNJ5:
Generating mutation plots.
Highly correlated genes for KCNJ5:
Showing top 20/861 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ5 | ABHD14B | 0.743575 | 3 | 0 | 3 |
| KCNJ5 | ZMAT4 | 0.743493 | 3 | 0 | 3 |
| KCNJ5 | UBR4 | 0.741809 | 3 | 0 | 3 |
| KCNJ5 | C2orf16 | 0.727356 | 5 | 0 | 4 |
| KCNJ5 | FAM71F2 | 0.726641 | 3 | 0 | 3 |
| KCNJ5 | ARGLU1 | 0.711022 | 3 | 0 | 3 |
| KCNJ5 | GRN | 0.702735 | 4 | 0 | 4 |
| KCNJ5 | SPRY1 | 0.701086 | 3 | 0 | 3 |
| KCNJ5 | PCAT4 | 0.696973 | 3 | 0 | 3 |
| KCNJ5 | MUC13 | 0.693311 | 3 | 0 | 3 |
| KCNJ5 | GORASP1 | 0.691092 | 4 | 0 | 4 |
| KCNJ5 | MTA3 | 0.688167 | 3 | 0 | 3 |
| KCNJ5 | GML | 0.684778 | 5 | 0 | 4 |
| KCNJ5 | MCHR2 | 0.683795 | 3 | 0 | 3 |
| KCNJ5 | MRPS18C | 0.682748 | 3 | 0 | 3 |
| KCNJ5 | BAGE | 0.680984 | 3 | 0 | 3 |
| KCNJ5 | CIDEC | 0.68037 | 3 | 0 | 3 |
| KCNJ5 | ZNF346 | 0.677479 | 3 | 0 | 3 |
| KCNJ5 | AKIRIN2 | 0.674505 | 3 | 0 | 3 |
| KCNJ5 | BCL2L1 | 0.672756 | 3 | 0 | 3 |
For details and further investigation, click here