| Full name: bestrophin 2 | Alias Symbol: FLJ20132 | ||
| Type: protein-coding gene | Cytoband: 19p13.13 | ||
| Entrez ID: 54831 | HGNC ID: HGNC:17107 | Ensembl Gene: ENSG00000039987 | OMIM ID: 607335 |
| Drug and gene relationship at DGIdb | |||
BEST2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04970 | Salivary secretion |
Expression of BEST2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BEST2 | 54831 | 207432_at | 0.2271 | 0.6051 | |
| GSE20347 | BEST2 | 54831 | 207432_at | 0.1238 | 0.2984 | |
| GSE23400 | BEST2 | 54831 | 207432_at | 0.0026 | 0.9738 | |
| GSE26886 | BEST2 | 54831 | 207432_at | 0.7117 | 0.0137 | |
| GSE29001 | BEST2 | 54831 | 207432_at | 0.2357 | 0.5441 | |
| GSE38129 | BEST2 | 54831 | 207432_at | 0.0117 | 0.9409 | |
| GSE45670 | BEST2 | 54831 | 207432_at | 0.3667 | 0.2420 | |
| GSE53622 | BEST2 | 54831 | 98808 | 0.1211 | 0.1184 | |
| GSE53624 | BEST2 | 54831 | 98808 | 0.3897 | 0.0000 | |
| GSE63941 | BEST2 | 54831 | 207432_at | 0.5379 | 0.0007 | |
| GSE77861 | BEST2 | 54831 | 207432_at | 0.1466 | 0.2765 | |
| GSE97050 | BEST2 | 54831 | A_23_P16225 | 0.0337 | 0.8928 | |
| TCGA | BEST2 | 54831 | RNAseq | 2.1724 | 0.0013 |
Upregulated datasets: 1; Downregulated datasets: 0.
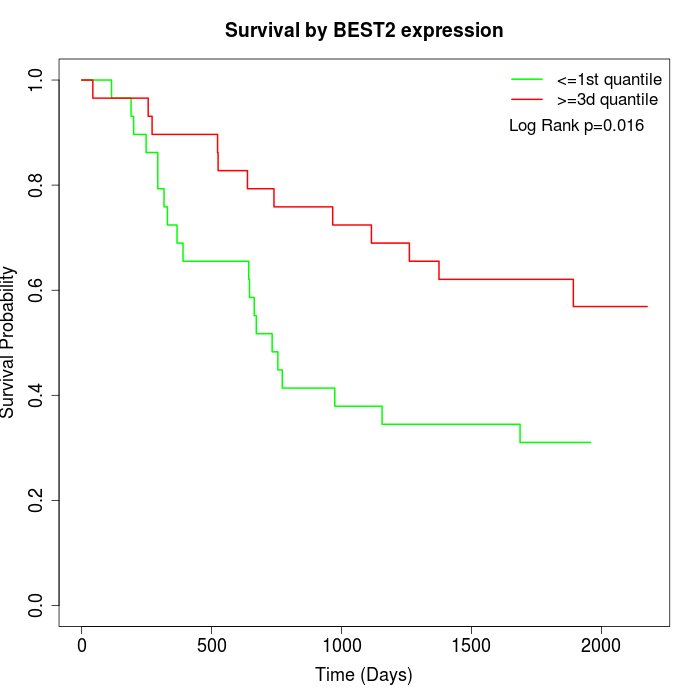
Survival by BEST2 expression:
Note: Click image to view full size file.
Copy number change of BEST2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BEST2 | 54831 | 4 | 2 | 24 | |
| GSE20123 | BEST2 | 54831 | 3 | 1 | 26 | |
| GSE43470 | BEST2 | 54831 | 2 | 6 | 35 | |
| GSE46452 | BEST2 | 54831 | 47 | 1 | 11 | |
| GSE47630 | BEST2 | 54831 | 4 | 7 | 29 | |
| GSE54993 | BEST2 | 54831 | 16 | 3 | 51 | |
| GSE54994 | BEST2 | 54831 | 6 | 13 | 34 | |
| GSE60625 | BEST2 | 54831 | 9 | 0 | 2 | |
| GSE74703 | BEST2 | 54831 | 2 | 4 | 30 | |
| GSE74704 | BEST2 | 54831 | 0 | 1 | 19 | |
| TCGA | BEST2 | 54831 | 16 | 13 | 67 |
Total number of gains: 109; Total number of losses: 51; Total Number of normals: 328.
Somatic mutations of BEST2:
Generating mutation plots.
Highly correlated genes for BEST2:
Showing top 20/141 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BEST2 | PIP5KL1 | 0.717251 | 3 | 0 | 3 |
| BEST2 | SDK1 | 0.701333 | 3 | 0 | 3 |
| BEST2 | SHC2 | 0.701169 | 3 | 0 | 3 |
| BEST2 | TOX2 | 0.695239 | 3 | 0 | 3 |
| BEST2 | FCRLB | 0.684791 | 3 | 0 | 3 |
| BEST2 | SNORA78 | 0.68258 | 3 | 0 | 3 |
| BEST2 | CRABP1 | 0.682161 | 4 | 0 | 4 |
| BEST2 | CDK16 | 0.679601 | 3 | 0 | 3 |
| BEST2 | PKM | 0.670415 | 3 | 0 | 3 |
| BEST2 | IL24 | 0.667574 | 3 | 0 | 3 |
| BEST2 | FOXA3 | 0.667006 | 3 | 0 | 3 |
| BEST2 | CFP | 0.665507 | 3 | 0 | 3 |
| BEST2 | FOXJ1 | 0.664692 | 4 | 0 | 3 |
| BEST2 | POC1A | 0.661718 | 4 | 0 | 3 |
| BEST2 | PROC | 0.657502 | 5 | 0 | 5 |
| BEST2 | HYAL1 | 0.655256 | 5 | 0 | 3 |
| BEST2 | RAP1GAP | 0.655186 | 3 | 0 | 3 |
| BEST2 | LRFN1 | 0.655126 | 4 | 0 | 3 |
| BEST2 | LYL1 | 0.653618 | 5 | 0 | 4 |
| BEST2 | BMF | 0.652932 | 4 | 0 | 3 |
For details and further investigation, click here