| Full name: RAP1 GTPase activating protein | Alias Symbol: KIAA0474|RAP1GAP1|RAP1GAPII | ||
| Type: protein-coding gene | Cytoband: 1p36.12 | ||
| Entrez ID: 5909 | HGNC ID: HGNC:9858 | Ensembl Gene: ENSG00000076864 | OMIM ID: 600278 |
| Drug and gene relationship at DGIdb | |||
RAP1GAP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway |
Expression of RAP1GAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAP1GAP | 5909 | 203911_at | -0.2169 | 0.6964 | |
| GSE20347 | RAP1GAP | 5909 | 203911_at | -0.0164 | 0.9440 | |
| GSE23400 | RAP1GAP | 5909 | 203911_at | -0.2049 | 0.0001 | |
| GSE26886 | RAP1GAP | 5909 | 203911_at | 0.3547 | 0.1884 | |
| GSE29001 | RAP1GAP | 5909 | 203911_at | -0.6233 | 0.0118 | |
| GSE38129 | RAP1GAP | 5909 | 203911_at | 0.0509 | 0.8478 | |
| GSE45670 | RAP1GAP | 5909 | 203911_at | 0.0019 | 0.9936 | |
| GSE53622 | RAP1GAP | 5909 | 146046 | -0.1171 | 0.4885 | |
| GSE53624 | RAP1GAP | 5909 | 146046 | -0.7683 | 0.0000 | |
| GSE63941 | RAP1GAP | 5909 | 203911_at | 1.1749 | 0.0864 | |
| GSE77861 | RAP1GAP | 5909 | 203911_at | -0.0195 | 0.9389 | |
| GSE97050 | RAP1GAP | 5909 | A_23_P45976 | -0.2992 | 0.2433 | |
| SRP007169 | RAP1GAP | 5909 | RNAseq | -1.2656 | 0.0306 | |
| SRP008496 | RAP1GAP | 5909 | RNAseq | -1.0350 | 0.0088 | |
| SRP064894 | RAP1GAP | 5909 | RNAseq | -1.6348 | 0.0000 | |
| SRP133303 | RAP1GAP | 5909 | RNAseq | -1.2123 | 0.0005 | |
| SRP159526 | RAP1GAP | 5909 | RNAseq | 1.0677 | 0.0671 | |
| SRP193095 | RAP1GAP | 5909 | RNAseq | 0.2172 | 0.5340 | |
| SRP219564 | RAP1GAP | 5909 | RNAseq | -0.7940 | 0.2605 | |
| TCGA | RAP1GAP | 5909 | RNAseq | -0.7951 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
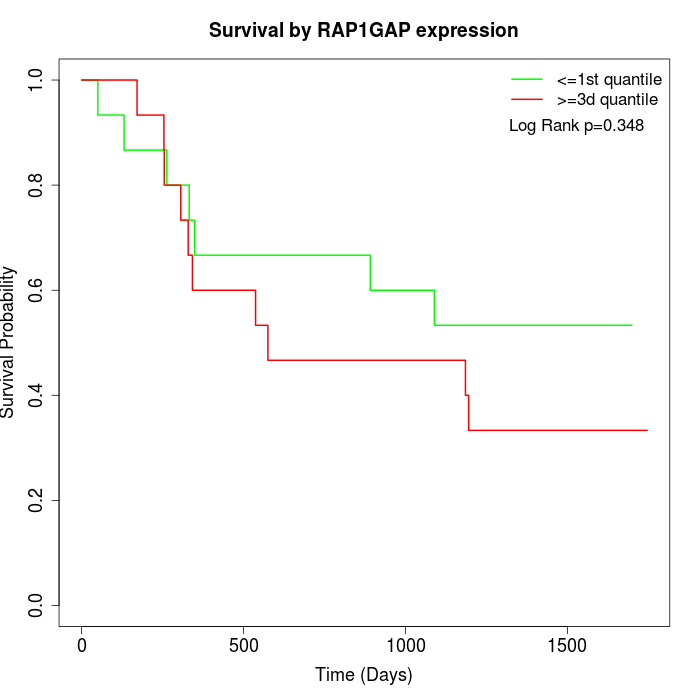
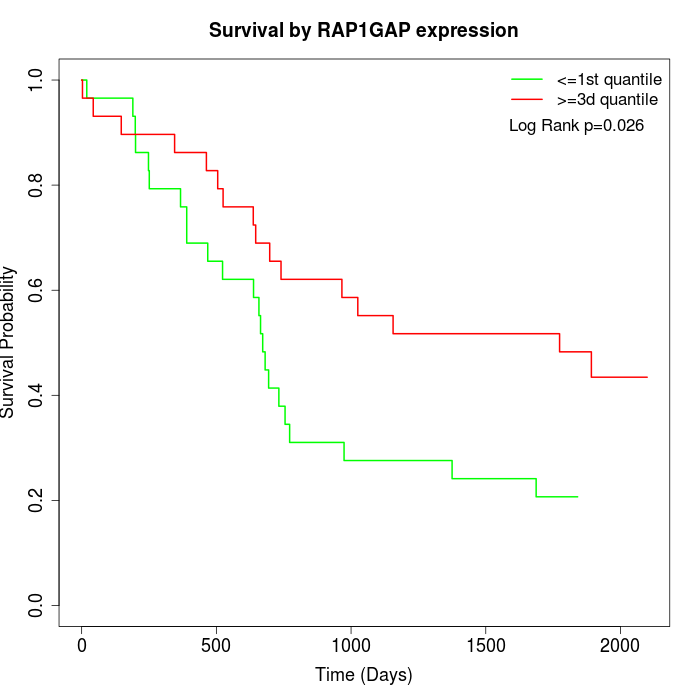
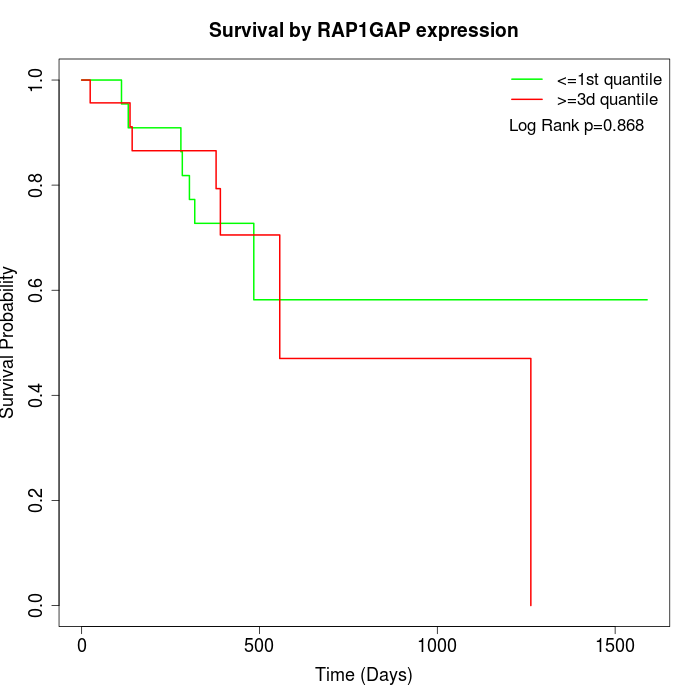
Survival by RAP1GAP expression:
Note: Click image to view full size file.
Copy number change of RAP1GAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAP1GAP | 5909 | 0 | 5 | 25 | |
| GSE20123 | RAP1GAP | 5909 | 0 | 4 | 26 | |
| GSE43470 | RAP1GAP | 5909 | 2 | 6 | 35 | |
| GSE46452 | RAP1GAP | 5909 | 5 | 1 | 53 | |
| GSE47630 | RAP1GAP | 5909 | 8 | 3 | 29 | |
| GSE54993 | RAP1GAP | 5909 | 3 | 1 | 66 | |
| GSE54994 | RAP1GAP | 5909 | 11 | 6 | 36 | |
| GSE60625 | RAP1GAP | 5909 | 0 | 0 | 11 | |
| GSE74703 | RAP1GAP | 5909 | 1 | 4 | 31 | |
| GSE74704 | RAP1GAP | 5909 | 0 | 0 | 20 | |
| TCGA | RAP1GAP | 5909 | 9 | 23 | 64 |
Total number of gains: 39; Total number of losses: 53; Total Number of normals: 396.
Somatic mutations of RAP1GAP:
Generating mutation plots.
Highly correlated genes for RAP1GAP:
Showing top 20/192 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAP1GAP | SULT2B1 | 0.696136 | 4 | 0 | 4 |
| RAP1GAP | WDR78 | 0.694961 | 3 | 0 | 3 |
| RAP1GAP | FAM223B | 0.681304 | 3 | 0 | 3 |
| RAP1GAP | CYP2D6 | 0.680532 | 3 | 0 | 3 |
| RAP1GAP | SYNPO2L | 0.679776 | 4 | 0 | 3 |
| RAP1GAP | TREH | 0.6795 | 5 | 0 | 5 |
| RAP1GAP | DEGS2 | 0.675369 | 3 | 0 | 3 |
| RAP1GAP | PMM1 | 0.664209 | 5 | 0 | 4 |
| RAP1GAP | SCAMP5 | 0.66117 | 3 | 0 | 3 |
| RAP1GAP | BEST2 | 0.655186 | 3 | 0 | 3 |
| RAP1GAP | LMOD3 | 0.651202 | 3 | 0 | 3 |
| RAP1GAP | GJB2 | 0.649173 | 3 | 0 | 3 |
| RAP1GAP | NANOS2 | 0.647878 | 5 | 0 | 5 |
| RAP1GAP | C1orf61 | 0.638354 | 3 | 0 | 3 |
| RAP1GAP | IQCH | 0.635626 | 4 | 0 | 3 |
| RAP1GAP | PYY | 0.635418 | 4 | 0 | 4 |
| RAP1GAP | CNNM1 | 0.635184 | 3 | 0 | 3 |
| RAP1GAP | ACSBG2 | 0.635152 | 3 | 0 | 3 |
| RAP1GAP | KCNK3 | 0.635098 | 3 | 0 | 3 |
| RAP1GAP | VSIG10L | 0.63267 | 4 | 0 | 4 |
For details and further investigation, click here