| Full name: BPI fold containing family A member 3 | Alias Symbol: bA49G10.4|SPLUNC3 | ||
| Type: protein-coding gene | Cytoband: 20q11.21 | ||
| Entrez ID: 128861 | HGNC ID: HGNC:16204 | Ensembl Gene: ENSG00000131059 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of BPIFA3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BPIFA3 | 128861 | 240359_at | 0.0136 | 0.9742 | |
| GSE26886 | BPIFA3 | 128861 | 240359_at | 0.0384 | 0.8263 | |
| GSE45670 | BPIFA3 | 128861 | 240359_at | -0.0336 | 0.7743 | |
| GSE53622 | BPIFA3 | 128861 | 88135 | 0.4611 | 0.0074 | |
| GSE53624 | BPIFA3 | 128861 | 88135 | 0.3167 | 0.0046 | |
| GSE63941 | BPIFA3 | 128861 | 240359_at | -0.1203 | 0.5371 | |
| GSE77861 | BPIFA3 | 128861 | 240359_at | -0.2049 | 0.0977 |
Upregulated datasets: 0; Downregulated datasets: 0.
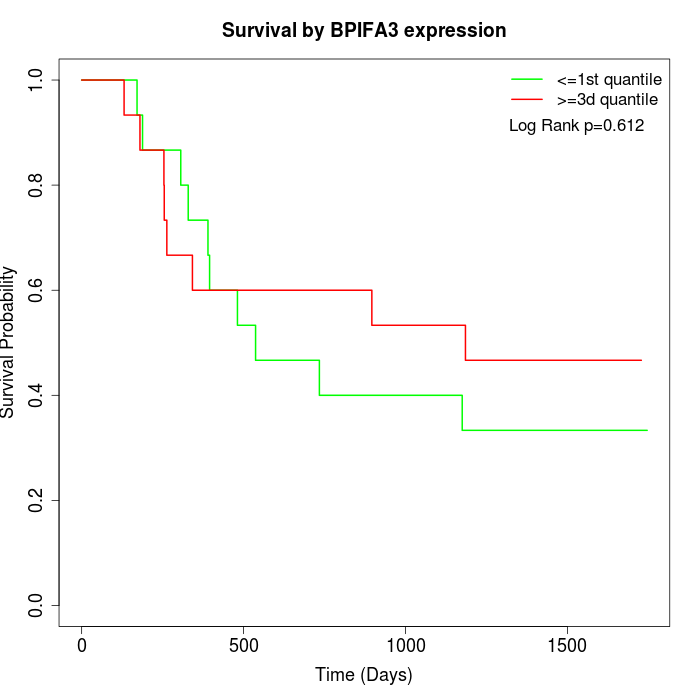
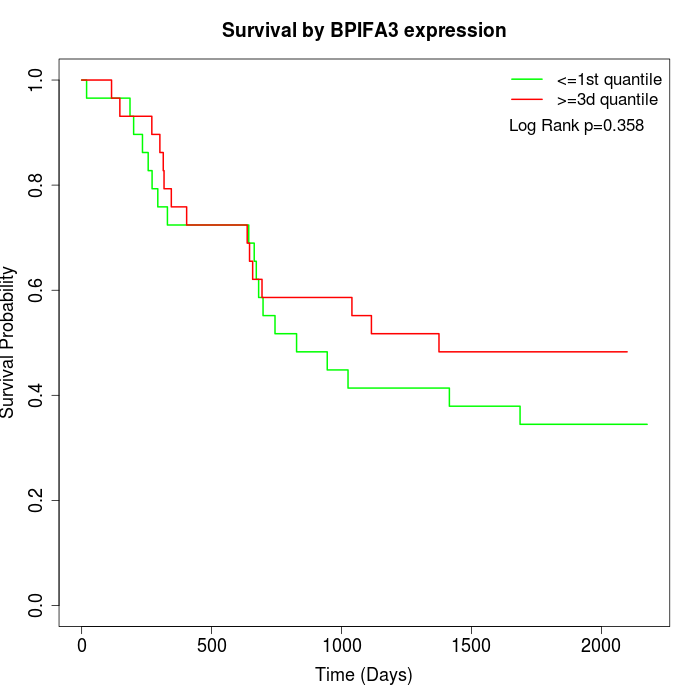
Survival by BPIFA3 expression:
Note: Click image to view full size file.
Copy number change of BPIFA3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BPIFA3 | 128861 | 15 | 0 | 15 | |
| GSE20123 | BPIFA3 | 128861 | 15 | 0 | 15 | |
| GSE43470 | BPIFA3 | 128861 | 12 | 0 | 31 | |
| GSE46452 | BPIFA3 | 128861 | 29 | 0 | 30 | |
| GSE47630 | BPIFA3 | 128861 | 24 | 1 | 15 | |
| GSE54993 | BPIFA3 | 128861 | 0 | 19 | 51 | |
| GSE54994 | BPIFA3 | 128861 | 26 | 1 | 26 | |
| GSE60625 | BPIFA3 | 128861 | 0 | 0 | 11 | |
| GSE74703 | BPIFA3 | 128861 | 11 | 0 | 25 | |
| GSE74704 | BPIFA3 | 128861 | 10 | 0 | 10 | |
| TCGA | BPIFA3 | 128861 | 50 | 1 | 45 |
Total number of gains: 192; Total number of losses: 22; Total Number of normals: 274.
Somatic mutations of BPIFA3:
Generating mutation plots.
Highly correlated genes for BPIFA3:
Showing top 20/77 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BPIFA3 | C10orf90 | 0.670941 | 3 | 0 | 3 |
| BPIFA3 | PRL | 0.658216 | 3 | 0 | 3 |
| BPIFA3 | CSH1 | 0.625994 | 3 | 0 | 3 |
| BPIFA3 | ABCC12 | 0.623121 | 3 | 0 | 3 |
| BPIFA3 | MYL2 | 0.622295 | 3 | 0 | 3 |
| BPIFA3 | HCN1 | 0.613831 | 3 | 0 | 3 |
| BPIFA3 | VGLL2 | 0.610132 | 3 | 0 | 3 |
| BPIFA3 | PTH2 | 0.605968 | 3 | 0 | 3 |
| BPIFA3 | FGF6 | 0.60505 | 3 | 0 | 3 |
| BPIFA3 | SLC5A2 | 0.597362 | 3 | 0 | 3 |
| BPIFA3 | CAMK2B | 0.59619 | 3 | 0 | 3 |
| BPIFA3 | DCST1 | 0.591247 | 4 | 0 | 3 |
| BPIFA3 | NHLRC4 | 0.591029 | 4 | 0 | 3 |
| BPIFA3 | CBLN4 | 0.588701 | 4 | 0 | 3 |
| BPIFA3 | ASB10 | 0.587359 | 4 | 0 | 3 |
| BPIFA3 | SLC6A19 | 0.586724 | 3 | 0 | 3 |
| BPIFA3 | OR10A3 | 0.58621 | 3 | 0 | 3 |
| BPIFA3 | MPV17L | 0.58605 | 3 | 0 | 3 |
| BPIFA3 | KRT37 | 0.58264 | 3 | 0 | 3 |
| BPIFA3 | TNFRSF13C | 0.57882 | 4 | 0 | 3 |
For details and further investigation, click here