| Full name: complement C6 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5p13.1 | ||
| Entrez ID: 729 | HGNC ID: HGNC:1339 | Ensembl Gene: ENSG00000039537 | OMIM ID: 217050 |
| Drug and gene relationship at DGIdb | |||
Expression of C6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C6 | 729 | 210168_at | -0.7543 | 0.2761 | |
| GSE20347 | C6 | 729 | 210168_at | -0.0702 | 0.4582 | |
| GSE23400 | C6 | 729 | 210168_at | -0.1623 | 0.0069 | |
| GSE26886 | C6 | 729 | 210168_at | -0.0343 | 0.8499 | |
| GSE29001 | C6 | 729 | 210168_at | -0.1914 | 0.3196 | |
| GSE38129 | C6 | 729 | 210168_at | -0.6440 | 0.0039 | |
| GSE45670 | C6 | 729 | 210168_at | -0.5641 | 0.0003 | |
| GSE53622 | C6 | 729 | 37040 | -1.3336 | 0.0000 | |
| GSE53624 | C6 | 729 | 37040 | -1.6799 | 0.0000 | |
| GSE63941 | C6 | 729 | 210168_at | 0.0608 | 0.7468 | |
| GSE77861 | C6 | 729 | 210168_at | 0.0160 | 0.9244 | |
| SRP133303 | C6 | 729 | RNAseq | -3.5201 | 0.0000 | |
| TCGA | C6 | 729 | RNAseq | -4.7095 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
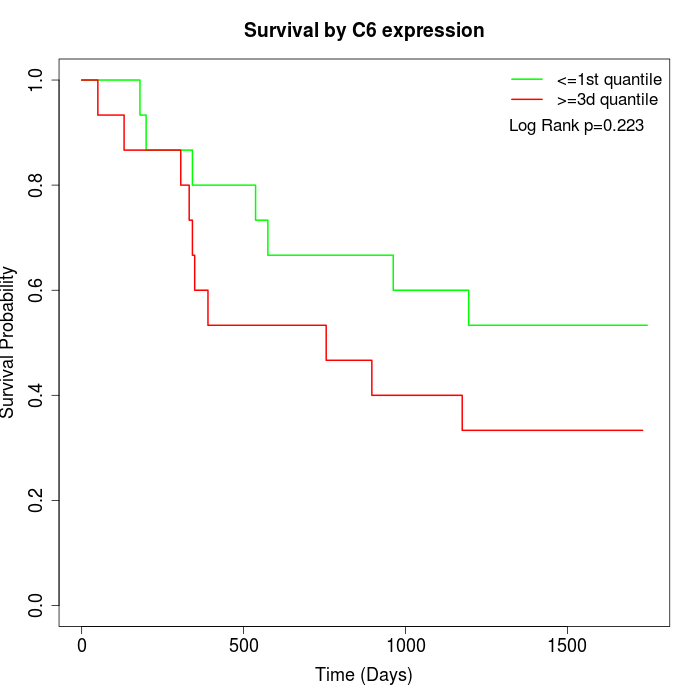
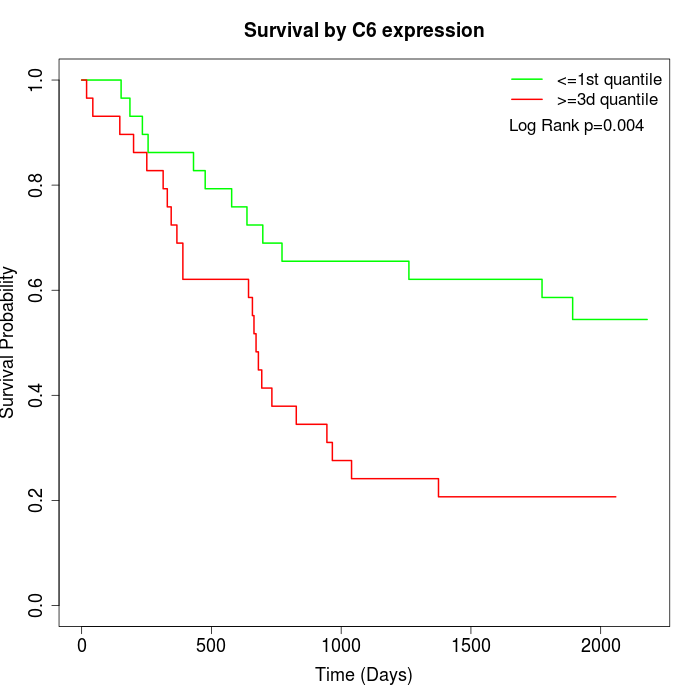
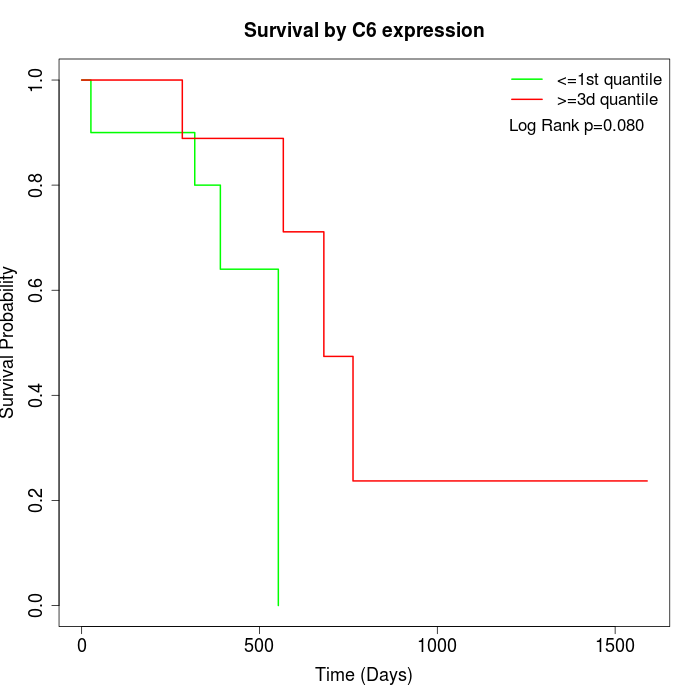
Survival by C6 expression:
Note: Click image to view full size file.
Copy number change of C6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C6 | 729 | 9 | 0 | 21 | |
| GSE20123 | C6 | 729 | 9 | 0 | 21 | |
| GSE43470 | C6 | 729 | 17 | 0 | 26 | |
| GSE46452 | C6 | 729 | 5 | 22 | 32 | |
| GSE47630 | C6 | 729 | 6 | 12 | 22 | |
| GSE54993 | C6 | 729 | 5 | 4 | 61 | |
| GSE54994 | C6 | 729 | 24 | 2 | 27 | |
| GSE60625 | C6 | 729 | 0 | 0 | 11 | |
| GSE74703 | C6 | 729 | 13 | 0 | 23 | |
| GSE74704 | C6 | 729 | 8 | 0 | 12 | |
| TCGA | C6 | 729 | 53 | 6 | 37 |
Total number of gains: 149; Total number of losses: 46; Total Number of normals: 293.
Somatic mutations of C6:
Generating mutation plots.
Highly correlated genes for C6:
Showing top 20/1284 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C6 | NPY1R | 0.753499 | 3 | 0 | 3 |
| C6 | GFRA4 | 0.744498 | 4 | 0 | 4 |
| C6 | PLCXD3 | 0.741569 | 5 | 0 | 5 |
| C6 | C1QTNF7 | 0.734388 | 5 | 0 | 5 |
| C6 | MAMDC2 | 0.732081 | 4 | 0 | 4 |
| C6 | NR6A1 | 0.720764 | 4 | 0 | 4 |
| C6 | INO80D | 0.719466 | 3 | 0 | 3 |
| C6 | CGA | 0.71881 | 3 | 0 | 3 |
| C6 | IFNA5 | 0.717824 | 3 | 0 | 3 |
| C6 | JAM2 | 0.715683 | 5 | 0 | 5 |
| C6 | EPX | 0.715567 | 4 | 0 | 4 |
| C6 | C8A | 0.715173 | 3 | 0 | 3 |
| C6 | MTFR1L | 0.713523 | 4 | 0 | 4 |
| C6 | MYOT | 0.712847 | 5 | 0 | 5 |
| C6 | LINC00889 | 0.709794 | 3 | 0 | 3 |
| C6 | NEGR1 | 0.70887 | 5 | 0 | 5 |
| C6 | PRR26 | 0.708112 | 5 | 0 | 5 |
| C6 | RNF150 | 0.706887 | 5 | 0 | 5 |
| C6 | VAT1L | 0.705062 | 4 | 0 | 4 |
| C6 | KCNJ10 | 0.702194 | 3 | 0 | 3 |
For details and further investigation, click here