| Full name: vesicle amine transport 1 like | Alias Symbol: KIAA1576 | ||
| Type: protein-coding gene | Cytoband: 16q23.1 | ||
| Entrez ID: 57687 | HGNC ID: HGNC:29315 | Ensembl Gene: ENSG00000171724 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of VAT1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VAT1L | 57687 | 226415_at | -0.1865 | 0.8104 | |
| GSE26886 | VAT1L | 57687 | 226415_at | 0.2375 | 0.1154 | |
| GSE45670 | VAT1L | 57687 | 226415_at | -0.5315 | 0.0579 | |
| GSE53622 | VAT1L | 57687 | 2130 | -0.7631 | 0.0002 | |
| GSE53624 | VAT1L | 57687 | 2130 | -0.8255 | 0.0000 | |
| GSE63941 | VAT1L | 57687 | 226415_at | -4.2360 | 0.0000 | |
| GSE77861 | VAT1L | 57687 | 226415_at | 0.0117 | 0.9419 | |
| SRP159526 | VAT1L | 57687 | RNAseq | -1.9529 | 0.0050 | |
| SRP219564 | VAT1L | 57687 | RNAseq | -1.9441 | 0.0144 | |
| TCGA | VAT1L | 57687 | RNAseq | -1.4329 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
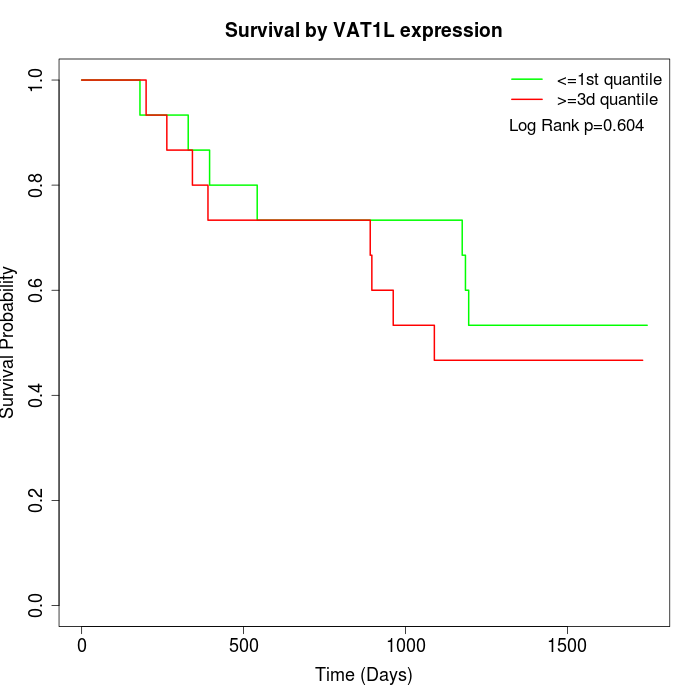
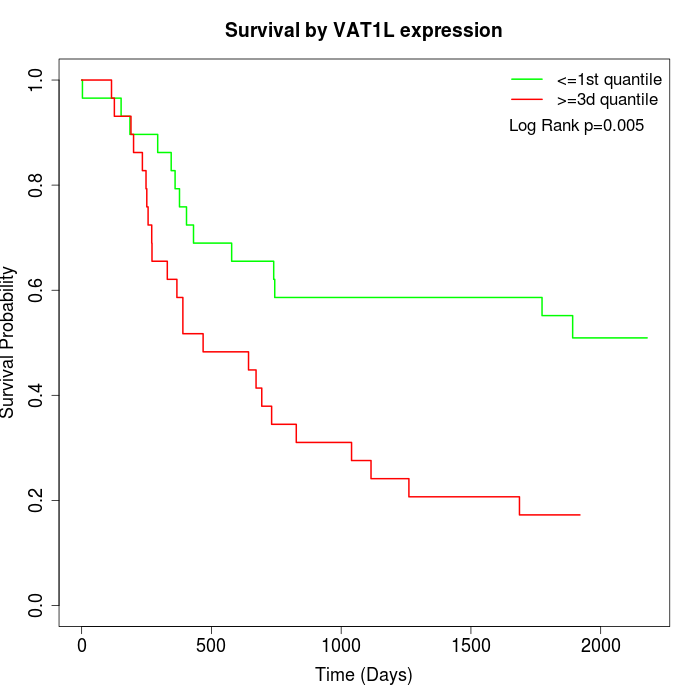
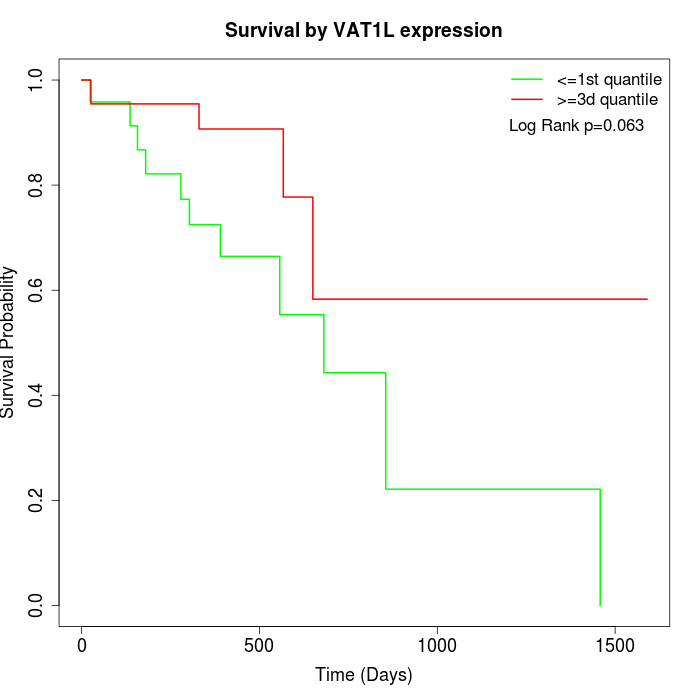
Survival by VAT1L expression:
Note: Click image to view full size file.
Copy number change of VAT1L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VAT1L | 57687 | 3 | 3 | 24 | |
| GSE20123 | VAT1L | 57687 | 3 | 3 | 24 | |
| GSE43470 | VAT1L | 57687 | 2 | 8 | 33 | |
| GSE46452 | VAT1L | 57687 | 37 | 2 | 20 | |
| GSE47630 | VAT1L | 57687 | 11 | 8 | 21 | |
| GSE54993 | VAT1L | 57687 | 2 | 4 | 64 | |
| GSE54994 | VAT1L | 57687 | 7 | 11 | 35 | |
| GSE60625 | VAT1L | 57687 | 4 | 0 | 7 | |
| GSE74703 | VAT1L | 57687 | 2 | 6 | 28 | |
| GSE74704 | VAT1L | 57687 | 2 | 2 | 16 | |
| TCGA | VAT1L | 57687 | 25 | 14 | 57 |
Total number of gains: 98; Total number of losses: 61; Total Number of normals: 329.
Somatic mutations of VAT1L:
Generating mutation plots.
Highly correlated genes for VAT1L:
Showing top 20/481 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VAT1L | DCHS1 | 0.744051 | 4 | 0 | 4 |
| VAT1L | ACO1 | 0.743119 | 3 | 0 | 3 |
| VAT1L | TCEAL7 | 0.723542 | 6 | 0 | 6 |
| VAT1L | DENND2A | 0.718966 | 5 | 0 | 5 |
| VAT1L | ZNF660 | 0.718177 | 5 | 0 | 5 |
| VAT1L | MCAM | 0.710648 | 4 | 0 | 4 |
| VAT1L | SYDE1 | 0.707042 | 5 | 0 | 4 |
| VAT1L | PCDH18 | 0.705923 | 4 | 0 | 4 |
| VAT1L | SGCD | 0.70568 | 6 | 0 | 6 |
| VAT1L | C6 | 0.705062 | 4 | 0 | 4 |
| VAT1L | TSC22D1-AS1 | 0.702013 | 3 | 0 | 3 |
| VAT1L | KIF5C | 0.697914 | 4 | 0 | 3 |
| VAT1L | FZD4 | 0.695312 | 5 | 0 | 5 |
| VAT1L | RHOQ | 0.6944 | 4 | 0 | 3 |
| VAT1L | DDR2 | 0.692653 | 6 | 0 | 6 |
| VAT1L | NME5 | 0.685211 | 5 | 0 | 5 |
| VAT1L | RGMB | 0.685133 | 4 | 0 | 3 |
| VAT1L | RUNX1T1 | 0.684244 | 4 | 0 | 4 |
| VAT1L | CNRIP1 | 0.683966 | 6 | 0 | 5 |
| VAT1L | LRCH2 | 0.683396 | 5 | 0 | 5 |
For details and further investigation, click here