| Full name: calcyphosine | Alias Symbol: CAPS1|MGC126562 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 828 | HGNC ID: HGNC:1487 | Ensembl Gene: ENSG00000105519 | OMIM ID: 114212 |
| Drug and gene relationship at DGIdb | |||
Expression of CAPS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CAPS | 828 | 226424_at | -0.2113 | 0.6027 | |
| GSE26886 | CAPS | 828 | 226424_at | 0.6432 | 0.0541 | |
| GSE45670 | CAPS | 828 | 226424_at | -0.1061 | 0.5110 | |
| GSE53622 | CAPS | 828 | 58096 | -0.6823 | 0.0000 | |
| GSE53624 | CAPS | 828 | 58096 | -1.2969 | 0.0000 | |
| GSE63941 | CAPS | 828 | 231729_s_at | 0.6605 | 0.3407 | |
| GSE77861 | CAPS | 828 | 226424_at | -0.0312 | 0.8318 | |
| GSE97050 | CAPS | 828 | A_24_P313993 | 0.1544 | 0.5695 | |
| SRP064894 | CAPS | 828 | RNAseq | 0.0152 | 0.9688 | |
| SRP133303 | CAPS | 828 | RNAseq | -0.5292 | 0.0500 | |
| SRP193095 | CAPS | 828 | RNAseq | -0.3149 | 0.1891 | |
| SRP219564 | CAPS | 828 | RNAseq | 0.1036 | 0.8496 | |
| TCGA | CAPS | 828 | RNAseq | -0.6617 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
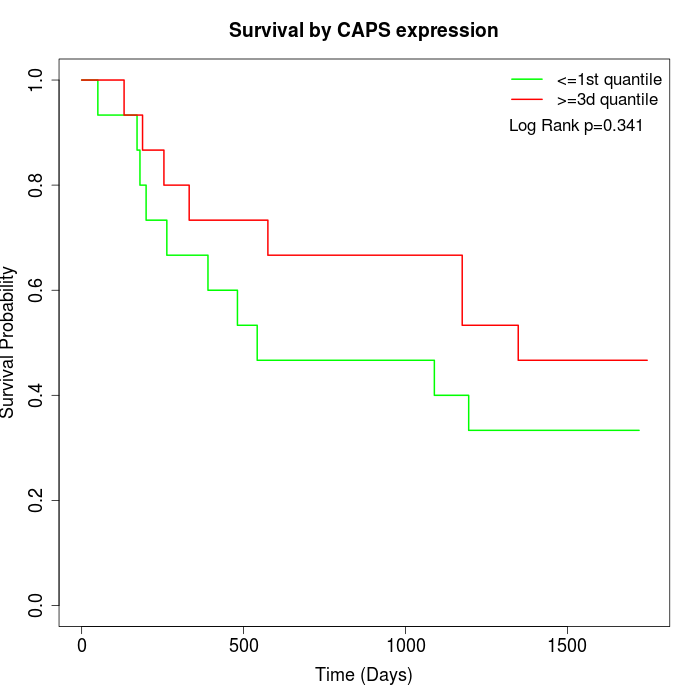
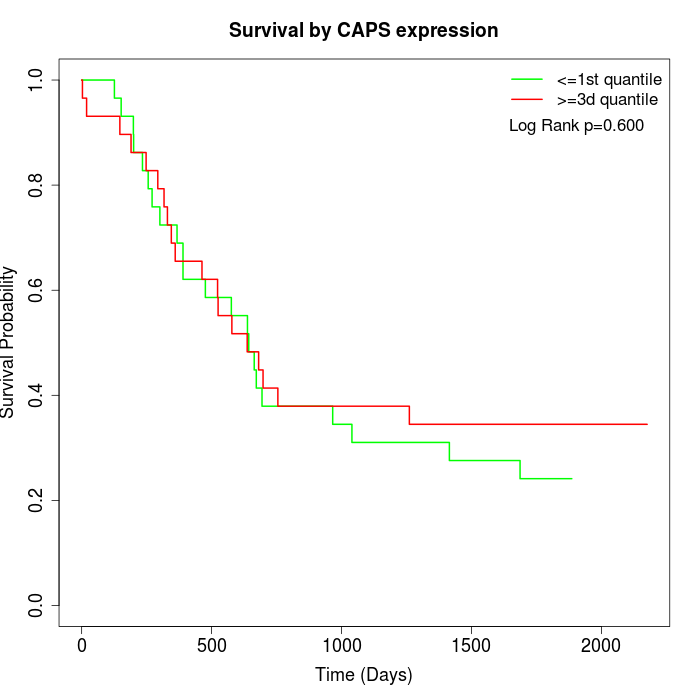
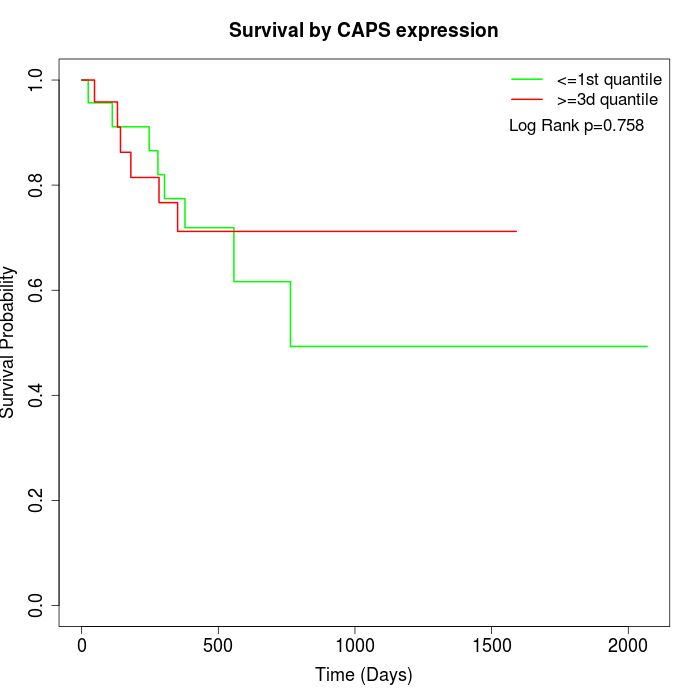
Survival by CAPS expression:
Note: Click image to view full size file.
Copy number change of CAPS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CAPS | 828 | 5 | 3 | 22 | |
| GSE20123 | CAPS | 828 | 4 | 2 | 24 | |
| GSE43470 | CAPS | 828 | 2 | 6 | 35 | |
| GSE46452 | CAPS | 828 | 47 | 1 | 11 | |
| GSE47630 | CAPS | 828 | 5 | 7 | 28 | |
| GSE54993 | CAPS | 828 | 16 | 3 | 51 | |
| GSE54994 | CAPS | 828 | 6 | 16 | 31 | |
| GSE60625 | CAPS | 828 | 9 | 0 | 2 | |
| GSE74703 | CAPS | 828 | 2 | 4 | 30 | |
| GSE74704 | CAPS | 828 | 1 | 2 | 17 | |
| TCGA | CAPS | 828 | 9 | 18 | 69 |
Total number of gains: 106; Total number of losses: 62; Total Number of normals: 320.
Somatic mutations of CAPS:
Generating mutation plots.
Highly correlated genes for CAPS:
Showing top 20/44 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CAPS | ELN | 0.644303 | 4 | 0 | 3 |
| CAPS | RALGPS1 | 0.643573 | 4 | 0 | 4 |
| CAPS | ZNF358 | 0.631422 | 4 | 0 | 4 |
| CAPS | MGAT3 | 0.618037 | 3 | 0 | 3 |
| CAPS | CEL | 0.616192 | 3 | 0 | 3 |
| CAPS | ATXN7L1 | 0.60688 | 3 | 0 | 3 |
| CAPS | EFHC2 | 0.605319 | 4 | 0 | 4 |
| CAPS | ZNF564 | 0.598584 | 3 | 0 | 3 |
| CAPS | KRT8 | 0.594851 | 3 | 0 | 3 |
| CAPS | PPFIBP2 | 0.592915 | 3 | 0 | 3 |
| CAPS | CCDC136 | 0.579372 | 4 | 0 | 3 |
| CAPS | PPM1L | 0.577649 | 4 | 0 | 3 |
| CAPS | CCDC28A | 0.573946 | 4 | 0 | 3 |
| CAPS | THBS4 | 0.571824 | 4 | 0 | 3 |
| CAPS | EFHD1 | 0.570607 | 4 | 0 | 3 |
| CAPS | NR3C2 | 0.56703 | 5 | 0 | 3 |
| CAPS | ZC3H10 | 0.56428 | 4 | 0 | 3 |
| CAPS | DPP7 | 0.561422 | 4 | 0 | 3 |
| CAPS | ARL1 | 0.560693 | 4 | 0 | 3 |
| CAPS | IDH3G | 0.559014 | 3 | 0 | 3 |
For details and further investigation, click here