| Full name: cancer susceptibility 22 | Alias Symbol: TCONS_00024290|LincRNA-ENST00000515084 | ||
| Type: non-coding RNA | Cytoband: 16q12.1 | ||
| Entrez ID: 283854 | HGNC ID: HGNC:50627 | Ensembl Gene: ENSG00000260887 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CASC22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CASC22 | 283854 | 1556954_at | -0.1776 | 0.4171 | |
| GSE26886 | CASC22 | 283854 | 1556954_at | 0.1028 | 0.2162 | |
| GSE45670 | CASC22 | 283854 | 1556954_at | 0.0339 | 0.6602 | |
| GSE53622 | CASC22 | 283854 | 104253 | 0.1300 | 0.1773 | |
| GSE53624 | CASC22 | 283854 | 104253 | 0.2315 | 0.0080 | |
| GSE63941 | CASC22 | 283854 | 1556954_at | 0.0065 | 0.9665 | |
| GSE77861 | CASC22 | 283854 | 1556954_at | -0.1224 | 0.3132 |
Upregulated datasets: 0; Downregulated datasets: 0.
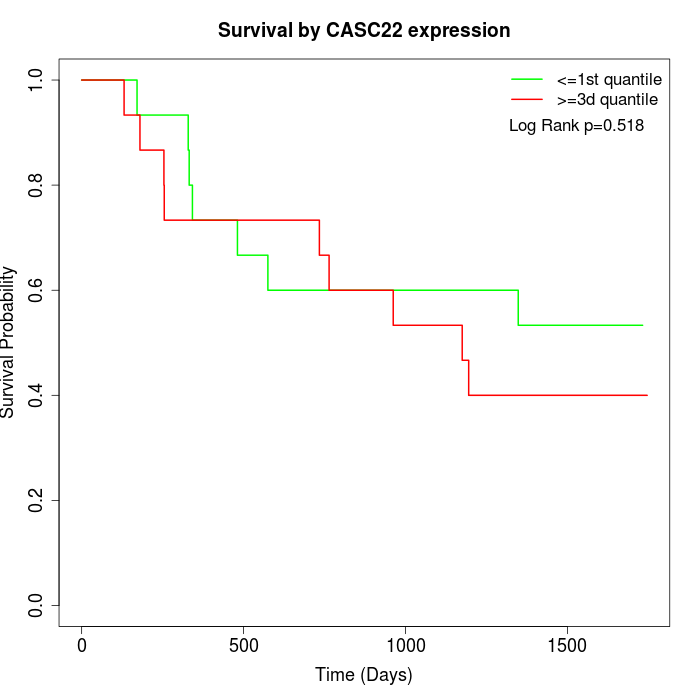
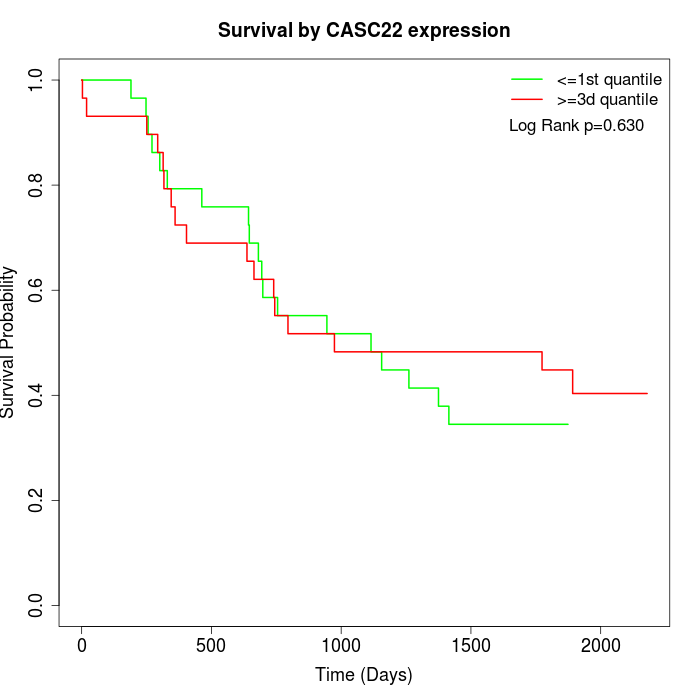
Survival by CASC22 expression:
Note: Click image to view full size file.
Copy number change of CASC22:
No record found for this gene.
Somatic mutations of CASC22:
Generating mutation plots.
Highly correlated genes for CASC22:
Showing top 20/56 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CASC22 | KCNF1 | 0.67916 | 3 | 0 | 3 |
| CASC22 | SLC4A9 | 0.67476 | 3 | 0 | 3 |
| CASC22 | NOTCH4 | 0.650568 | 3 | 0 | 3 |
| CASC22 | PKLR | 0.634563 | 4 | 0 | 4 |
| CASC22 | NPHS2 | 0.633735 | 3 | 0 | 3 |
| CASC22 | CDH22 | 0.631085 | 3 | 0 | 3 |
| CASC22 | APOC3 | 0.617746 | 3 | 0 | 3 |
| CASC22 | SEZ6 | 0.60747 | 3 | 0 | 3 |
| CASC22 | AGRP | 0.603391 | 3 | 0 | 3 |
| CASC22 | ANKRD33 | 0.598368 | 4 | 0 | 3 |
| CASC22 | NPPB | 0.597691 | 4 | 0 | 3 |
| CASC22 | MC3R | 0.596772 | 4 | 0 | 3 |
| CASC22 | CDHR5 | 0.593678 | 4 | 0 | 3 |
| CASC22 | APC2 | 0.590964 | 3 | 0 | 3 |
| CASC22 | CACNG7 | 0.590946 | 5 | 0 | 3 |
| CASC22 | KCNA7 | 0.586513 | 3 | 0 | 3 |
| CASC22 | CCDC154 | 0.585397 | 4 | 0 | 3 |
| CASC22 | SSMEM1 | 0.58441 | 3 | 0 | 3 |
| CASC22 | SCN2B | 0.584309 | 4 | 0 | 3 |
| CASC22 | HMCN2 | 0.580568 | 4 | 0 | 3 |
For details and further investigation, click here