| Full name: C-C motif chemokine ligand 2 | Alias Symbol: MCP1|MCP-1|MCAF|SMC-CF|GDCF-2|HC11|MGC9434 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 6347 | HGNC ID: HGNC:10618 | Ensembl Gene: ENSG00000108691 | OMIM ID: 158105 |
| Drug and gene relationship at DGIdb | |||
CCL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications |
Expression of CCL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL2 | 6347 | 216598_s_at | -0.6548 | 0.6880 | |
| GSE20347 | CCL2 | 6347 | 216598_s_at | 0.1878 | 0.6723 | |
| GSE23400 | CCL2 | 6347 | 216598_s_at | -0.1139 | 0.4867 | |
| GSE26886 | CCL2 | 6347 | 216598_s_at | 0.6332 | 0.0524 | |
| GSE29001 | CCL2 | 6347 | 216598_s_at | -0.0101 | 0.9855 | |
| GSE38129 | CCL2 | 6347 | 216598_s_at | -0.2720 | 0.5795 | |
| GSE45670 | CCL2 | 6347 | 216598_s_at | -1.7219 | 0.0034 | |
| GSE53622 | CCL2 | 6347 | 49592 | -0.2658 | 0.2273 | |
| GSE53624 | CCL2 | 6347 | 49592 | -0.1628 | 0.3786 | |
| GSE63941 | CCL2 | 6347 | 216598_s_at | -7.4506 | 0.0000 | |
| GSE77861 | CCL2 | 6347 | 216598_s_at | 0.3740 | 0.2849 | |
| GSE97050 | CCL2 | 6347 | A_23_P89431 | 0.1142 | 0.8536 | |
| SRP007169 | CCL2 | 6347 | RNAseq | 1.4447 | 0.0065 | |
| SRP008496 | CCL2 | 6347 | RNAseq | 2.4906 | 0.0000 | |
| SRP064894 | CCL2 | 6347 | RNAseq | -0.6073 | 0.1969 | |
| SRP133303 | CCL2 | 6347 | RNAseq | 0.3002 | 0.3275 | |
| SRP159526 | CCL2 | 6347 | RNAseq | 0.1336 | 0.8306 | |
| SRP193095 | CCL2 | 6347 | RNAseq | 1.7991 | 0.0000 | |
| SRP219564 | CCL2 | 6347 | RNAseq | -1.1240 | 0.2851 | |
| TCGA | CCL2 | 6347 | RNAseq | -0.0500 | 0.7825 |
Upregulated datasets: 3; Downregulated datasets: 2.
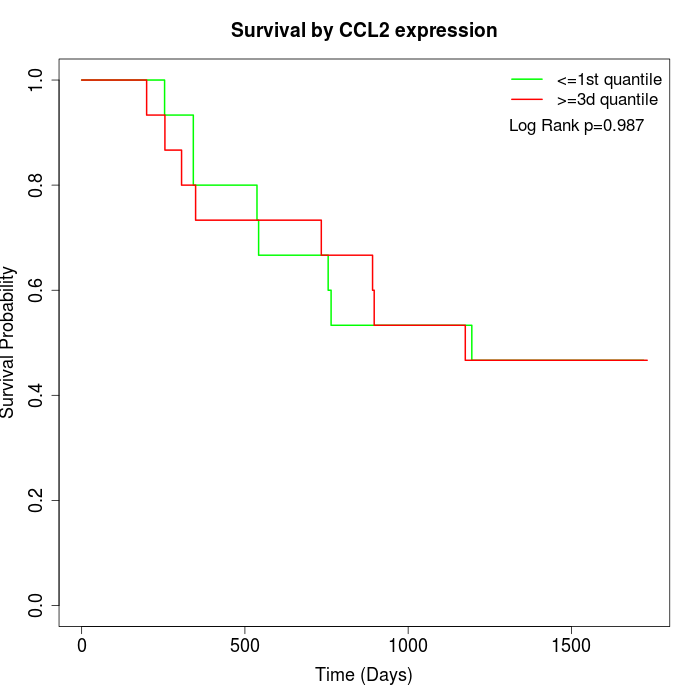
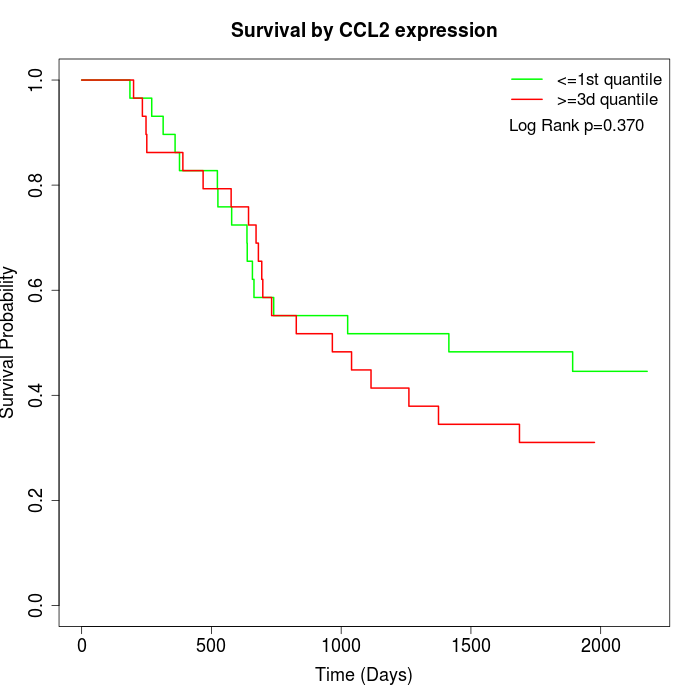
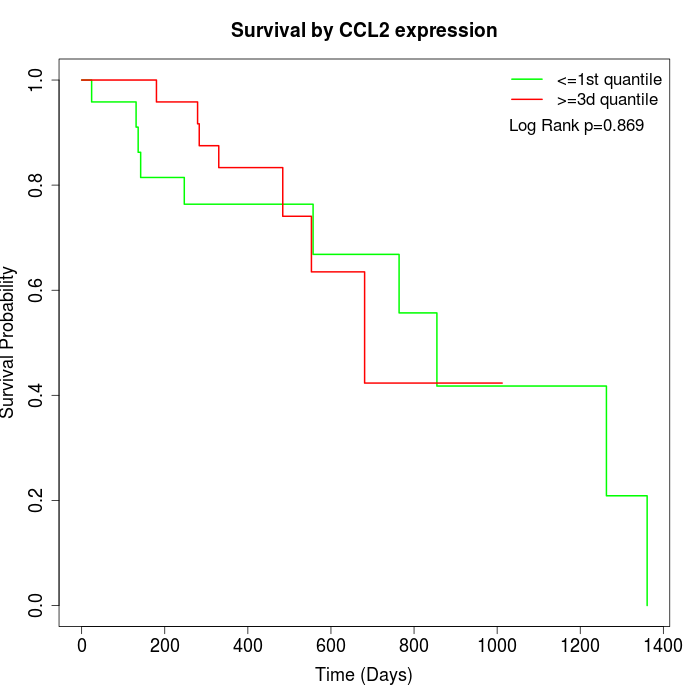
Survival by CCL2 expression:
Note: Click image to view full size file.
Copy number change of CCL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL2 | 6347 | 5 | 1 | 24 | |
| GSE20123 | CCL2 | 6347 | 5 | 1 | 24 | |
| GSE43470 | CCL2 | 6347 | 1 | 2 | 40 | |
| GSE46452 | CCL2 | 6347 | 34 | 0 | 25 | |
| GSE47630 | CCL2 | 6347 | 7 | 1 | 32 | |
| GSE54993 | CCL2 | 6347 | 3 | 3 | 64 | |
| GSE54994 | CCL2 | 6347 | 8 | 6 | 39 | |
| GSE60625 | CCL2 | 6347 | 4 | 0 | 7 | |
| GSE74703 | CCL2 | 6347 | 1 | 1 | 34 | |
| GSE74704 | CCL2 | 6347 | 3 | 1 | 16 | |
| TCGA | CCL2 | 6347 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of CCL2:
Generating mutation plots.
Highly correlated genes for CCL2:
Showing top 20/752 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL2 | C11orf96 | 0.819542 | 4 | 0 | 4 |
| CCL2 | SPATA9 | 0.776382 | 3 | 0 | 3 |
| CCL2 | MEDAG | 0.753762 | 4 | 0 | 4 |
| CCL2 | IL6 | 0.749484 | 9 | 0 | 9 |
| CCL2 | DPY19L1 | 0.749075 | 3 | 0 | 3 |
| CCL2 | SYTL4 | 0.734747 | 3 | 0 | 3 |
| CCL2 | ZNF791 | 0.732272 | 3 | 0 | 3 |
| CCL2 | FGF3 | 0.714449 | 3 | 0 | 3 |
| CCL2 | SMPD1 | 0.713374 | 4 | 0 | 3 |
| CCL2 | DLC1 | 0.713236 | 8 | 0 | 8 |
| CCL2 | PRPH2 | 0.689043 | 5 | 0 | 5 |
| CCL2 | TMEM140 | 0.688343 | 5 | 0 | 5 |
| CCL2 | SLC39A13 | 0.685491 | 4 | 0 | 3 |
| CCL2 | ANXA6 | 0.684997 | 10 | 0 | 10 |
| CCL2 | DPP4 | 0.682077 | 4 | 0 | 3 |
| CCL2 | WTIP | 0.681584 | 5 | 0 | 4 |
| CCL2 | TBC1D8B | 0.680524 | 3 | 0 | 3 |
| CCL2 | GNPAT | 0.679696 | 3 | 0 | 3 |
| CCL2 | SYNC | 0.676347 | 5 | 0 | 4 |
| CCL2 | GALNT10 | 0.674484 | 4 | 0 | 3 |
For details and further investigation, click here