| Full name: glyceronephosphate O-acyltransferase | Alias Symbol: DHAPAT|DAPAT|DAP-AT | ||
| Type: protein-coding gene | Cytoband: 1q42.2 | ||
| Entrez ID: 8443 | HGNC ID: HGNC:4416 | Ensembl Gene: ENSG00000116906 | OMIM ID: 602744 |
| Drug and gene relationship at DGIdb | |||
Expression of GNPAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNPAT | 8443 | 201956_s_at | 0.2627 | 0.3626 | |
| GSE20347 | GNPAT | 8443 | 201956_s_at | 0.2919 | 0.0071 | |
| GSE23400 | GNPAT | 8443 | 201956_s_at | 0.2709 | 0.0000 | |
| GSE26886 | GNPAT | 8443 | 201956_s_at | 0.6786 | 0.0004 | |
| GSE29001 | GNPAT | 8443 | 201956_s_at | 0.1576 | 0.6977 | |
| GSE38129 | GNPAT | 8443 | 201956_s_at | 0.1065 | 0.2773 | |
| GSE45670 | GNPAT | 8443 | 201956_s_at | 0.0919 | 0.3132 | |
| GSE53622 | GNPAT | 8443 | 103451 | 0.2840 | 0.0000 | |
| GSE53624 | GNPAT | 8443 | 103451 | 0.3170 | 0.0000 | |
| GSE63941 | GNPAT | 8443 | 201956_s_at | -0.1332 | 0.7432 | |
| GSE77861 | GNPAT | 8443 | 201956_s_at | 0.2982 | 0.1069 | |
| GSE97050 | GNPAT | 8443 | A_23_P85777 | -0.1990 | 0.5010 | |
| SRP007169 | GNPAT | 8443 | RNAseq | 0.5261 | 0.1842 | |
| SRP008496 | GNPAT | 8443 | RNAseq | 0.5430 | 0.0531 | |
| SRP064894 | GNPAT | 8443 | RNAseq | 0.1602 | 0.1689 | |
| SRP133303 | GNPAT | 8443 | RNAseq | 0.1898 | 0.0862 | |
| SRP159526 | GNPAT | 8443 | RNAseq | 0.1371 | 0.6544 | |
| SRP193095 | GNPAT | 8443 | RNAseq | -0.1105 | 0.2757 | |
| SRP219564 | GNPAT | 8443 | RNAseq | -0.0567 | 0.8517 | |
| TCGA | GNPAT | 8443 | RNAseq | -0.0837 | 0.0726 |
Upregulated datasets: 0; Downregulated datasets: 0.
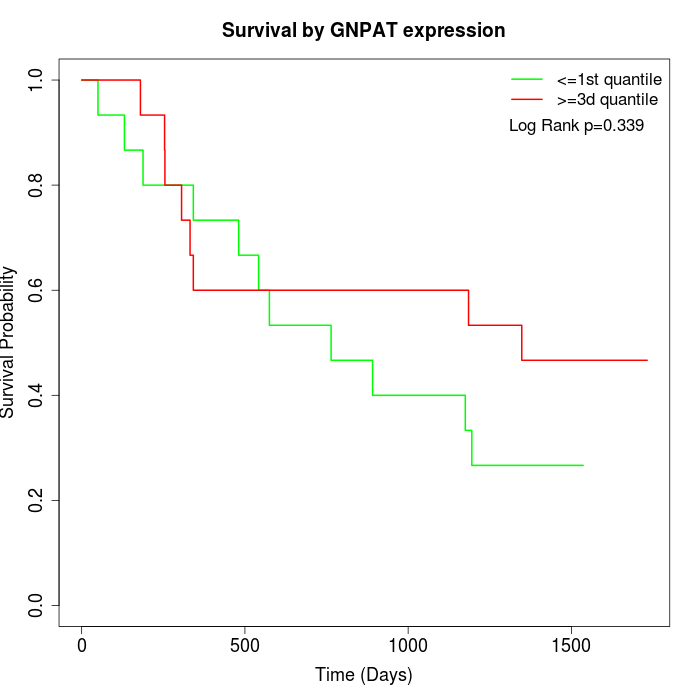
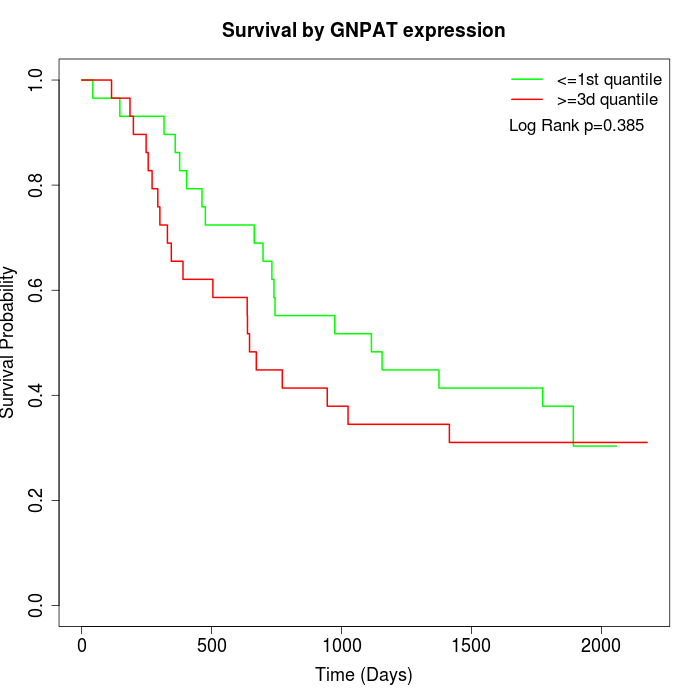
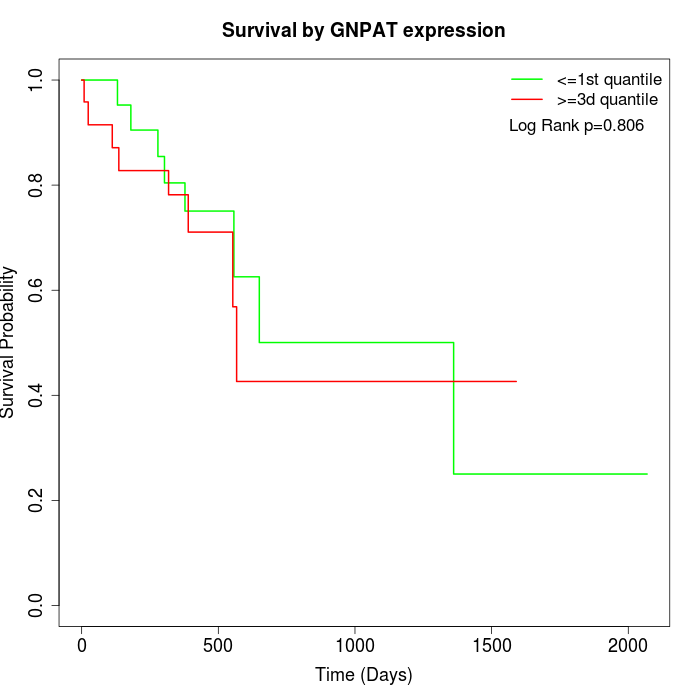
Survival by GNPAT expression:
Note: Click image to view full size file.
Copy number change of GNPAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNPAT | 8443 | 12 | 0 | 18 | |
| GSE20123 | GNPAT | 8443 | 12 | 0 | 18 | |
| GSE43470 | GNPAT | 8443 | 8 | 1 | 34 | |
| GSE46452 | GNPAT | 8443 | 4 | 2 | 53 | |
| GSE47630 | GNPAT | 8443 | 15 | 0 | 25 | |
| GSE54993 | GNPAT | 8443 | 0 | 6 | 64 | |
| GSE54994 | GNPAT | 8443 | 17 | 0 | 36 | |
| GSE60625 | GNPAT | 8443 | 0 | 0 | 11 | |
| GSE74703 | GNPAT | 8443 | 8 | 1 | 27 | |
| GSE74704 | GNPAT | 8443 | 6 | 0 | 14 | |
| TCGA | GNPAT | 8443 | 44 | 4 | 48 |
Total number of gains: 126; Total number of losses: 14; Total Number of normals: 348.
Somatic mutations of GNPAT:
Generating mutation plots.
Highly correlated genes for GNPAT:
Showing top 20/1125 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNPAT | SPEN | 0.796005 | 3 | 0 | 3 |
| GNPAT | EIF3L | 0.781378 | 3 | 0 | 3 |
| GNPAT | LTBR | 0.770755 | 3 | 0 | 3 |
| GNPAT | KCTD12 | 0.76967 | 3 | 0 | 3 |
| GNPAT | TMEM25 | 0.758894 | 3 | 0 | 3 |
| GNPAT | MRPS31 | 0.757769 | 3 | 0 | 3 |
| GNPAT | PPP1R35 | 0.745974 | 3 | 0 | 3 |
| GNPAT | PSMG3 | 0.742539 | 3 | 0 | 3 |
| GNPAT | SLCO3A1 | 0.737979 | 3 | 0 | 3 |
| GNPAT | RPL19 | 0.735682 | 3 | 0 | 3 |
| GNPAT | IDS | 0.72894 | 3 | 0 | 3 |
| GNPAT | PNMA1 | 0.728564 | 3 | 0 | 3 |
| GNPAT | ANAPC4 | 0.724711 | 3 | 0 | 3 |
| GNPAT | ERVW-1 | 0.718999 | 3 | 0 | 3 |
| GNPAT | RPA2 | 0.717129 | 3 | 0 | 3 |
| GNPAT | SFXN4 | 0.716129 | 3 | 0 | 3 |
| GNPAT | SYNM | 0.712653 | 3 | 0 | 3 |
| GNPAT | RPP38 | 0.711658 | 3 | 0 | 3 |
| GNPAT | CCDC127 | 0.711493 | 3 | 0 | 3 |
| GNPAT | PPRC1 | 0.711452 | 4 | 0 | 3 |
For details and further investigation, click here