| Full name: mesenteric estrogen dependent adipogenesis | Alias Symbol: FLJ14834|AWMS3|MEDA-4 | ||
| Type: protein-coding gene | Cytoband: 13q12.3 | ||
| Entrez ID: 84935 | HGNC ID: HGNC:25926 | Ensembl Gene: ENSG00000102802 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of MEDAG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MEDAG | 84935 | 227058_at | -0.7678 | 0.4185 | |
| GSE26886 | MEDAG | 84935 | 227058_at | 0.0481 | 0.8820 | |
| GSE45670 | MEDAG | 84935 | 1565765_x_at | -0.0608 | 0.3870 | |
| GSE53622 | MEDAG | 84935 | 12377 | -0.3137 | 0.1228 | |
| GSE53624 | MEDAG | 84935 | 12377 | 0.0571 | 0.6964 | |
| GSE63941 | MEDAG | 84935 | 227058_at | -5.3775 | 0.0000 | |
| GSE77861 | MEDAG | 84935 | 227058_at | -0.0428 | 0.8106 | |
| SRP007169 | MEDAG | 84935 | RNAseq | 2.9417 | 0.0000 | |
| SRP008496 | MEDAG | 84935 | RNAseq | 4.1430 | 0.0000 | |
| SRP064894 | MEDAG | 84935 | RNAseq | 0.5386 | 0.3527 | |
| SRP133303 | MEDAG | 84935 | RNAseq | -0.3187 | 0.4631 | |
| SRP159526 | MEDAG | 84935 | RNAseq | 0.0818 | 0.8855 | |
| SRP219564 | MEDAG | 84935 | RNAseq | 1.0105 | 0.2837 |
Upregulated datasets: 2; Downregulated datasets: 1.
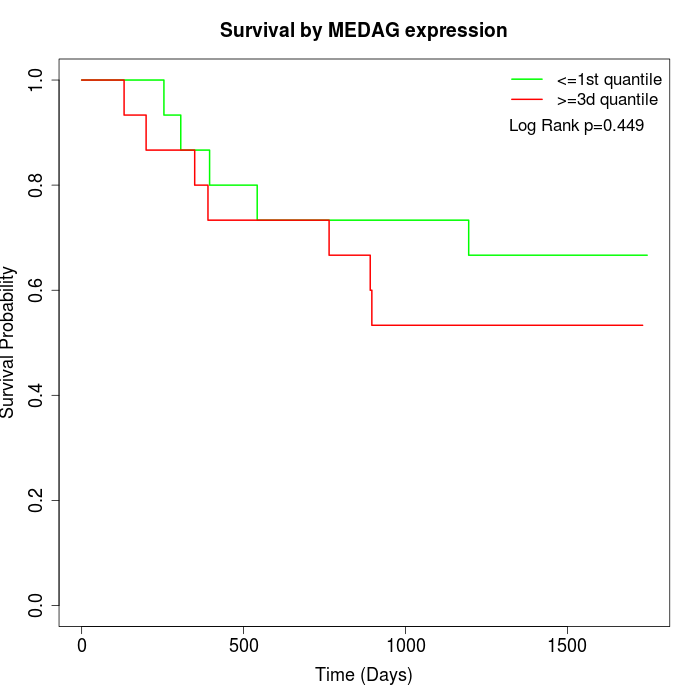
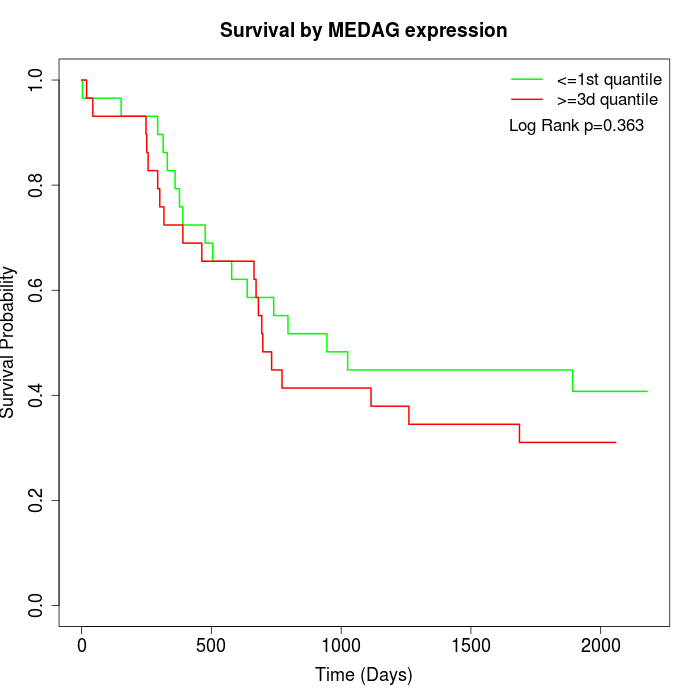
Survival by MEDAG expression:
Note: Click image to view full size file.
Copy number change of MEDAG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MEDAG | 84935 | 1 | 14 | 15 | |
| GSE20123 | MEDAG | 84935 | 1 | 13 | 16 | |
| GSE43470 | MEDAG | 84935 | 2 | 12 | 29 | |
| GSE46452 | MEDAG | 84935 | 0 | 33 | 26 | |
| GSE47630 | MEDAG | 84935 | 2 | 27 | 11 | |
| GSE54993 | MEDAG | 84935 | 12 | 2 | 56 | |
| GSE54994 | MEDAG | 84935 | 2 | 15 | 36 | |
| GSE60625 | MEDAG | 84935 | 0 | 3 | 8 | |
| GSE74703 | MEDAG | 84935 | 2 | 9 | 25 | |
| GSE74704 | MEDAG | 84935 | 0 | 11 | 9 | |
| TCGA | MEDAG | 84935 | 8 | 37 | 51 |
Total number of gains: 30; Total number of losses: 176; Total Number of normals: 282.
Somatic mutations of MEDAG:
Generating mutation plots.
Highly correlated genes for MEDAG:
Showing top 20/91 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MEDAG | CCL2 | 0.753762 | 4 | 0 | 4 |
| MEDAG | CXCL12 | 0.676966 | 5 | 0 | 4 |
| MEDAG | COL15A1 | 0.67415 | 5 | 0 | 5 |
| MEDAG | GSTM5 | 0.673598 | 4 | 0 | 4 |
| MEDAG | COLEC10 | 0.672667 | 3 | 0 | 3 |
| MEDAG | ARHGAP31 | 0.667987 | 3 | 0 | 3 |
| MEDAG | CRISPLD2 | 0.658445 | 5 | 0 | 4 |
| MEDAG | LINC01007 | 0.654505 | 3 | 0 | 3 |
| MEDAG | FAT4 | 0.654441 | 4 | 0 | 4 |
| MEDAG | ACOT12 | 0.649915 | 4 | 0 | 4 |
| MEDAG | NR4A3 | 0.647706 | 6 | 0 | 5 |
| MEDAG | DCN | 0.646225 | 5 | 0 | 5 |
| MEDAG | CNRIP1 | 0.645409 | 5 | 0 | 5 |
| MEDAG | THBS1 | 0.64315 | 5 | 0 | 5 |
| MEDAG | FIBIN | 0.639754 | 4 | 0 | 3 |
| MEDAG | DGKI | 0.635966 | 4 | 0 | 3 |
| MEDAG | FAM8A1 | 0.634742 | 3 | 0 | 3 |
| MEDAG | RASSF8-AS1 | 0.632699 | 3 | 0 | 3 |
| MEDAG | CHRM5 | 0.629251 | 3 | 0 | 3 |
| MEDAG | CDK15 | 0.629176 | 4 | 0 | 3 |
For details and further investigation, click here