| Full name: cyclin D2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.32 | ||
| Entrez ID: 894 | HGNC ID: HGNC:1583 | Ensembl Gene: ENSG00000118971 | OMIM ID: 123833 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CCND2 involved pathways:
Expression of CCND2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCND2 | 894 | 200953_s_at | -1.3534 | 0.1159 | |
| GSE20347 | CCND2 | 894 | 200953_s_at | -0.6319 | 0.0889 | |
| GSE23400 | CCND2 | 894 | 200953_s_at | -0.3799 | 0.0768 | |
| GSE26886 | CCND2 | 894 | 200953_s_at | -0.5786 | 0.2497 | |
| GSE29001 | CCND2 | 894 | 200953_s_at | 0.0689 | 0.9317 | |
| GSE38129 | CCND2 | 894 | 200953_s_at | -1.0320 | 0.0008 | |
| GSE45670 | CCND2 | 894 | 200953_s_at | -0.2966 | 0.4952 | |
| GSE53622 | CCND2 | 894 | 154986 | -0.6306 | 0.0000 | |
| GSE53624 | CCND2 | 894 | 154986 | -0.6058 | 0.0000 | |
| GSE63941 | CCND2 | 894 | 200953_s_at | -2.5590 | 0.2814 | |
| GSE77861 | CCND2 | 894 | 200953_s_at | -0.1885 | 0.9003 | |
| GSE97050 | CCND2 | 894 | A_24_P278747 | -0.6579 | 0.1186 | |
| SRP007169 | CCND2 | 894 | RNAseq | 2.4179 | 0.0001 | |
| SRP008496 | CCND2 | 894 | RNAseq | 2.4429 | 0.0000 | |
| SRP064894 | CCND2 | 894 | RNAseq | -0.5629 | 0.0967 | |
| SRP133303 | CCND2 | 894 | RNAseq | -0.2284 | 0.4742 | |
| SRP159526 | CCND2 | 894 | RNAseq | -0.1141 | 0.8196 | |
| SRP193095 | CCND2 | 894 | RNAseq | 0.1063 | 0.7155 | |
| SRP219564 | CCND2 | 894 | RNAseq | -0.4853 | 0.3474 | |
| TCGA | CCND2 | 894 | RNAseq | -0.2270 | 0.0867 |
Upregulated datasets: 2; Downregulated datasets: 1.
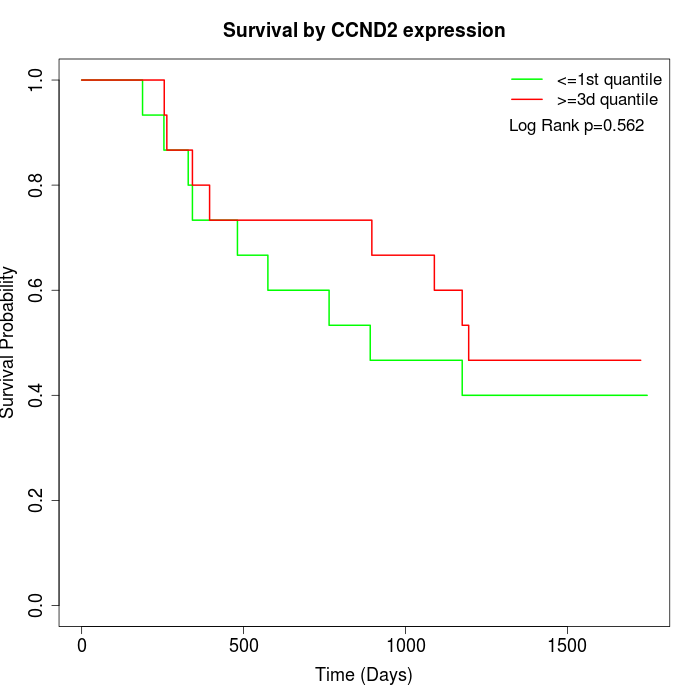
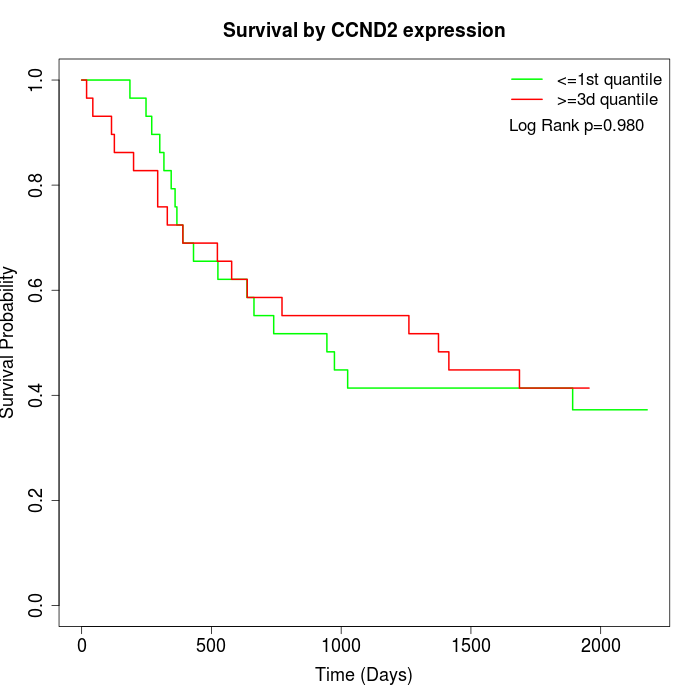
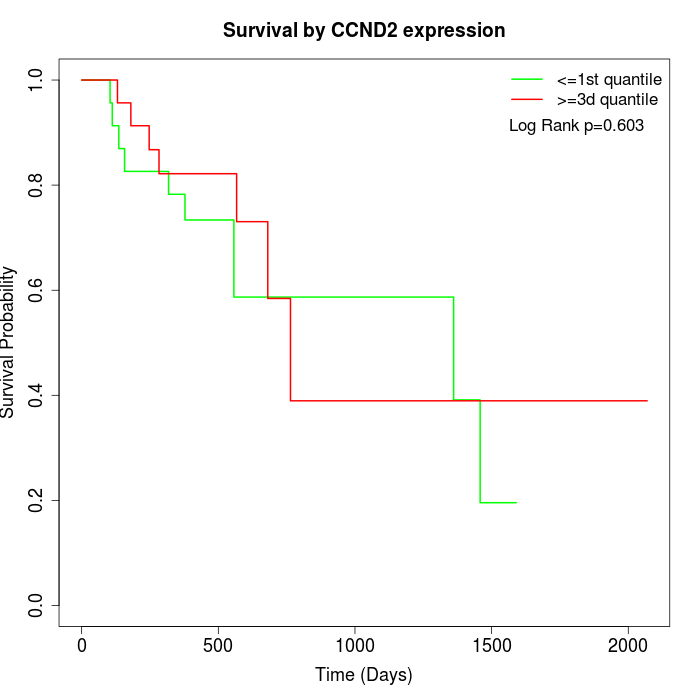
Survival by CCND2 expression:
Note: Click image to view full size file.
Copy number change of CCND2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCND2 | 894 | 6 | 4 | 20 | |
| GSE20123 | CCND2 | 894 | 6 | 4 | 20 | |
| GSE43470 | CCND2 | 894 | 10 | 3 | 30 | |
| GSE46452 | CCND2 | 894 | 10 | 1 | 48 | |
| GSE47630 | CCND2 | 894 | 12 | 2 | 26 | |
| GSE54993 | CCND2 | 894 | 1 | 10 | 59 | |
| GSE54994 | CCND2 | 894 | 10 | 2 | 41 | |
| GSE60625 | CCND2 | 894 | 0 | 1 | 10 | |
| GSE74703 | CCND2 | 894 | 9 | 2 | 25 | |
| GSE74704 | CCND2 | 894 | 4 | 2 | 14 | |
| TCGA | CCND2 | 894 | 40 | 6 | 50 |
Total number of gains: 108; Total number of losses: 37; Total Number of normals: 343.
Somatic mutations of CCND2:
Generating mutation plots.
Highly correlated genes for CCND2:
Showing top 20/457 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCND2 | MRVI1 | 0.7385 | 4 | 0 | 4 |
| CCND2 | EMCN | 0.721682 | 3 | 0 | 3 |
| CCND2 | SMPD1 | 0.719243 | 3 | 0 | 3 |
| CCND2 | MACROD2 | 0.718217 | 3 | 0 | 3 |
| CCND2 | CACNA1C | 0.692379 | 3 | 0 | 3 |
| CCND2 | RASSF3 | 0.688828 | 3 | 0 | 3 |
| CCND2 | HABP4 | 0.68231 | 3 | 0 | 3 |
| CCND2 | KIAA0408 | 0.679498 | 3 | 0 | 3 |
| CCND2 | PTGER3 | 0.679303 | 4 | 0 | 4 |
| CCND2 | PREX2 | 0.672631 | 4 | 0 | 4 |
| CCND2 | PPP3CB | 0.666147 | 5 | 0 | 4 |
| CCND2 | HOXA2 | 0.662916 | 4 | 0 | 3 |
| CCND2 | BAG3 | 0.659417 | 4 | 0 | 3 |
| CCND2 | LRRN4CL | 0.659371 | 4 | 0 | 4 |
| CCND2 | PHYHIPL | 0.657689 | 3 | 0 | 3 |
| CCND2 | NEXN | 0.656428 | 4 | 0 | 4 |
| CCND2 | PDLIM3 | 0.655721 | 6 | 0 | 6 |
| CCND2 | SLC2A12 | 0.655388 | 3 | 0 | 3 |
| CCND2 | SLIT3 | 0.653865 | 4 | 0 | 3 |
| CCND2 | MTRF1L | 0.649278 | 3 | 0 | 3 |
For details and further investigation, click here