| Full name: prostaglandin E receptor 3 | Alias Symbol: EP3 | ||
| Type: protein-coding gene | Cytoband: 1p31.1 | ||
| Entrez ID: 5733 | HGNC ID: HGNC:9595 | Ensembl Gene: ENSG00000050628 | OMIM ID: 176806 |
| Drug and gene relationship at DGIdb | |||
PTGER3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04024 | cAMP signaling pathway | |
| hsa04923 | Regulation of lipolysis in adipocytes |
Expression of PTGER3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PTGER3 | 5733 | 210374_x_at | -0.3095 | 0.1557 | |
| GSE20347 | PTGER3 | 5733 | 210832_x_at | 0.0212 | 0.8524 | |
| GSE23400 | PTGER3 | 5733 | 210832_x_at | -0.1162 | 0.0004 | |
| GSE26886 | PTGER3 | 5733 | 210374_x_at | -0.0249 | 0.8923 | |
| GSE29001 | PTGER3 | 5733 | 210832_x_at | -0.0501 | 0.8121 | |
| GSE38129 | PTGER3 | 5733 | 213933_at | -0.1352 | 0.6917 | |
| GSE45670 | PTGER3 | 5733 | 210374_x_at | -0.4475 | 0.0001 | |
| GSE53622 | PTGER3 | 5733 | 133391 | 0.8432 | 0.0000 | |
| GSE53624 | PTGER3 | 5733 | 133391 | 0.9019 | 0.0000 | |
| GSE63941 | PTGER3 | 5733 | 210374_x_at | -0.5276 | 0.0109 | |
| GSE77861 | PTGER3 | 5733 | 211909_x_at | -0.1623 | 0.4473 | |
| GSE97050 | PTGER3 | 5733 | A_33_P3265749 | 0.1140 | 0.9160 | |
| SRP007169 | PTGER3 | 5733 | RNAseq | 2.7626 | 0.0001 | |
| SRP008496 | PTGER3 | 5733 | RNAseq | 2.5422 | 0.0000 | |
| SRP064894 | PTGER3 | 5733 | RNAseq | 0.5801 | 0.0679 | |
| SRP133303 | PTGER3 | 5733 | RNAseq | 0.5845 | 0.0203 | |
| SRP159526 | PTGER3 | 5733 | RNAseq | 0.4405 | 0.4291 | |
| SRP193095 | PTGER3 | 5733 | RNAseq | -0.1439 | 0.6998 | |
| SRP219564 | PTGER3 | 5733 | RNAseq | -0.3599 | 0.7176 | |
| TCGA | PTGER3 | 5733 | RNAseq | -1.4315 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 1.
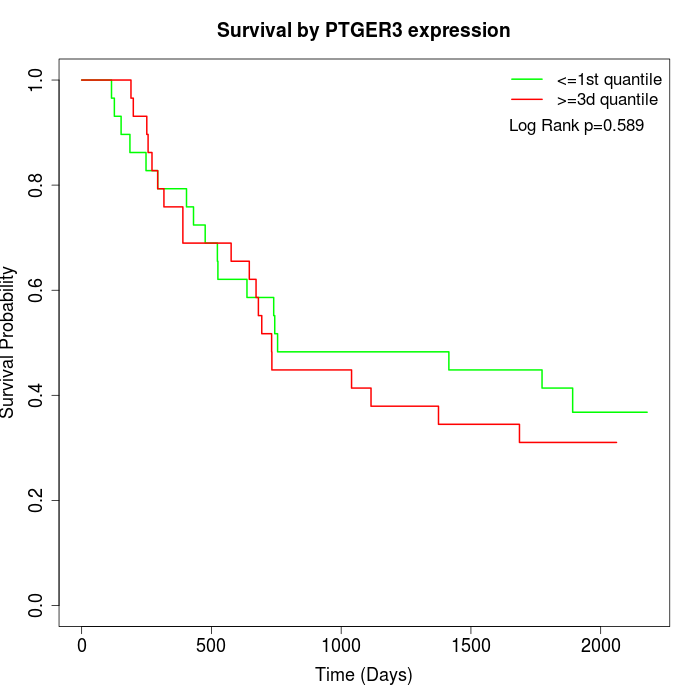
Survival by PTGER3 expression:
Note: Click image to view full size file.
Copy number change of PTGER3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PTGER3 | 5733 | 1 | 6 | 23 | |
| GSE20123 | PTGER3 | 5733 | 1 | 6 | 23 | |
| GSE43470 | PTGER3 | 5733 | 4 | 4 | 35 | |
| GSE46452 | PTGER3 | 5733 | 0 | 1 | 58 | |
| GSE47630 | PTGER3 | 5733 | 7 | 6 | 27 | |
| GSE54993 | PTGER3 | 5733 | 0 | 1 | 69 | |
| GSE54994 | PTGER3 | 5733 | 6 | 5 | 42 | |
| GSE60625 | PTGER3 | 5733 | 0 | 0 | 11 | |
| GSE74703 | PTGER3 | 5733 | 3 | 3 | 30 | |
| GSE74704 | PTGER3 | 5733 | 1 | 3 | 16 | |
| TCGA | PTGER3 | 5733 | 9 | 22 | 65 |
Total number of gains: 32; Total number of losses: 57; Total Number of normals: 399.
Somatic mutations of PTGER3:
Generating mutation plots.
Highly correlated genes for PTGER3:
Showing top 20/766 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PTGER3 | DMRTA1 | 0.741539 | 3 | 0 | 3 |
| PTGER3 | HMCN2 | 0.738595 | 3 | 0 | 3 |
| PTGER3 | CBR4 | 0.736633 | 3 | 0 | 3 |
| PTGER3 | KDM5D | 0.728265 | 3 | 0 | 3 |
| PTGER3 | DDX3Y | 0.725019 | 3 | 0 | 3 |
| PTGER3 | ZNF709 | 0.723999 | 3 | 0 | 3 |
| PTGER3 | ITGA11 | 0.722856 | 3 | 0 | 3 |
| PTGER3 | GULP1 | 0.718736 | 3 | 0 | 3 |
| PTGER3 | ARRDC3 | 0.714976 | 3 | 0 | 3 |
| PTGER3 | SPRY1 | 0.711908 | 4 | 0 | 4 |
| PTGER3 | RNF14 | 0.70844 | 3 | 0 | 3 |
| PTGER3 | ACO1 | 0.707072 | 4 | 0 | 4 |
| PTGER3 | LYSMD3 | 0.705517 | 3 | 0 | 3 |
| PTGER3 | SARDH | 0.702737 | 5 | 0 | 5 |
| PTGER3 | ZNF358 | 0.698313 | 4 | 0 | 3 |
| PTGER3 | GDF6 | 0.695239 | 3 | 0 | 3 |
| PTGER3 | DCHS1 | 0.689819 | 5 | 0 | 4 |
| PTGER3 | ZEB2 | 0.68867 | 5 | 0 | 4 |
| PTGER3 | IL33 | 0.68698 | 4 | 0 | 4 |
| PTGER3 | DIAPH2 | 0.684226 | 3 | 0 | 3 |
For details and further investigation, click here