| Full name: C-C motif chemokine receptor 2 | Alias Symbol: CC-CKR-2|CKR2|MCP-1-R|CD192|FLJ78302 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 729230 | HGNC ID: HGNC:1603 | Ensembl Gene: ENSG00000121807 | OMIM ID: 601267 |
| Drug and gene relationship at DGIdb | |||
CCR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCR2 | 729230 | 206978_at | -0.0686 | 0.9519 | |
| GSE20347 | CCR2 | 729230 | 206978_at | 0.0183 | 0.8854 | |
| GSE23400 | CCR2 | 729230 | 206978_at | -0.0733 | 0.1494 | |
| GSE26886 | CCR2 | 729230 | 206978_at | -0.8673 | 0.0060 | |
| GSE29001 | CCR2 | 729230 | 206978_at | 0.0195 | 0.9176 | |
| GSE38129 | CCR2 | 729230 | 206978_at | 0.0990 | 0.6310 | |
| GSE45670 | CCR2 | 729230 | 206978_at | -0.4822 | 0.1408 | |
| GSE53622 | CCR2 | 729230 | 21092 | -0.7538 | 0.0002 | |
| GSE53624 | CCR2 | 729230 | 23785 | -0.4752 | 0.1037 | |
| GSE63941 | CCR2 | 729230 | 206978_at | 0.0392 | 0.8096 | |
| GSE77861 | CCR2 | 729230 | 206978_at | -0.1978 | 0.1408 | |
| GSE97050 | CCR2 | 729230 | A_23_P324885 | 0.8143 | 0.2763 | |
| SRP007169 | CCR2 | 729230 | RNAseq | 0.9706 | 0.2434 | |
| SRP064894 | CCR2 | 729230 | RNAseq | 0.5194 | 0.1601 | |
| SRP133303 | CCR2 | 729230 | RNAseq | -0.6947 | 0.0062 | |
| SRP193095 | CCR2 | 729230 | RNAseq | -0.5215 | 0.0726 | |
| SRP219564 | CCR2 | 729230 | RNAseq | 0.3167 | 0.6404 | |
| TCGA | CCR2 | 729230 | RNAseq | -0.8946 | 0.0157 |
Upregulated datasets: 0; Downregulated datasets: 0.
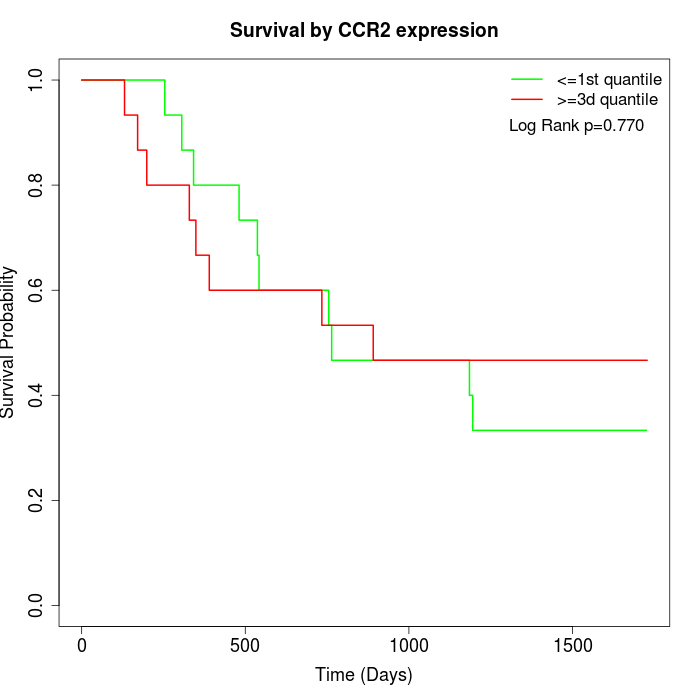
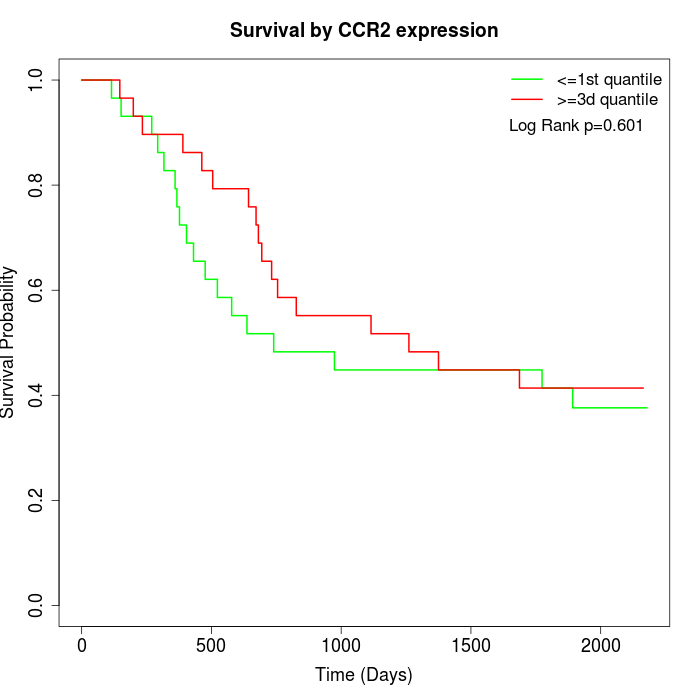
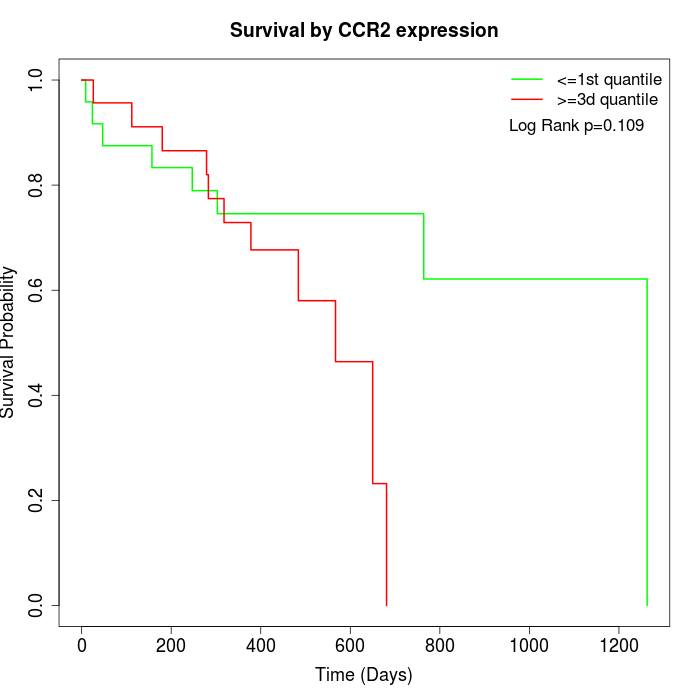
Survival by CCR2 expression:
Note: Click image to view full size file.
Copy number change of CCR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCR2 | 729230 | 0 | 17 | 13 | |
| GSE20123 | CCR2 | 729230 | 0 | 18 | 12 | |
| GSE43470 | CCR2 | 729230 | 0 | 20 | 23 | |
| GSE46452 | CCR2 | 729230 | 2 | 17 | 40 | |
| GSE47630 | CCR2 | 729230 | 2 | 23 | 15 | |
| GSE54993 | CCR2 | 729230 | 7 | 2 | 61 | |
| GSE54994 | CCR2 | 729230 | 0 | 36 | 17 | |
| GSE60625 | CCR2 | 729230 | 5 | 0 | 6 | |
| GSE74703 | CCR2 | 729230 | 0 | 15 | 21 | |
| GSE74704 | CCR2 | 729230 | 0 | 12 | 8 | |
| TCGA | CCR2 | 729230 | 1 | 75 | 20 |
Total number of gains: 17; Total number of losses: 235; Total Number of normals: 236.
Somatic mutations of CCR2:
Generating mutation plots.
Highly correlated genes for CCR2:
Showing top 20/501 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCR2 | MPEG1 | 0.796913 | 6 | 0 | 6 |
| CCR2 | GIMAP7 | 0.793061 | 6 | 0 | 6 |
| CCR2 | RGS18 | 0.783009 | 6 | 0 | 6 |
| CCR2 | C19orf38 | 0.757474 | 3 | 0 | 3 |
| CCR2 | GAB3 | 0.757198 | 5 | 0 | 5 |
| CCR2 | SLAMF6 | 0.753138 | 4 | 0 | 4 |
| CCR2 | ARHGAP15 | 0.752693 | 3 | 0 | 3 |
| CCR2 | GAPT | 0.75249 | 6 | 0 | 6 |
| CCR2 | C1orf162 | 0.746772 | 6 | 0 | 5 |
| CCR2 | GIMAP6 | 0.729266 | 9 | 0 | 7 |
| CCR2 | SDHB | 0.724022 | 3 | 0 | 3 |
| CCR2 | TNFAIP8L2 | 0.720063 | 6 | 0 | 5 |
| CCR2 | P2RY8 | 0.718322 | 4 | 0 | 3 |
| CCR2 | FLI1 | 0.716342 | 8 | 0 | 6 |
| CCR2 | GIMAP5 | 0.713508 | 4 | 0 | 3 |
| CCR2 | NOA1 | 0.711735 | 4 | 0 | 4 |
| CCR2 | ZNF90 | 0.708456 | 3 | 0 | 3 |
| CCR2 | SCML4 | 0.707729 | 4 | 0 | 4 |
| CCR2 | SUSD3 | 0.703744 | 5 | 0 | 5 |
| CCR2 | PDE4B | 0.703616 | 4 | 0 | 3 |
For details and further investigation, click here