| Full name: phosphodiesterase 4B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p31.3 | ||
| Entrez ID: 5142 | HGNC ID: HGNC:8781 | Ensembl Gene: ENSG00000184588 | OMIM ID: 600127 |
| Drug and gene relationship at DGIdb | |||
PDE4B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway |
Expression of PDE4B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE4B | 5142 | 203708_at | -0.0465 | 0.9798 | |
| GSE20347 | PDE4B | 5142 | 203708_at | 0.4311 | 0.3088 | |
| GSE23400 | PDE4B | 5142 | 211302_s_at | 0.1653 | 0.0629 | |
| GSE26886 | PDE4B | 5142 | 203708_at | 0.7107 | 0.1857 | |
| GSE29001 | PDE4B | 5142 | 203708_at | 0.5087 | 0.4333 | |
| GSE38129 | PDE4B | 5142 | 203708_at | 0.2123 | 0.5884 | |
| GSE45670 | PDE4B | 5142 | 203708_at | -0.6902 | 0.1636 | |
| GSE53622 | PDE4B | 5142 | 17859 | 0.2289 | 0.2254 | |
| GSE53624 | PDE4B | 5142 | 17859 | -0.0578 | 0.7595 | |
| GSE63941 | PDE4B | 5142 | 203708_at | -3.8750 | 0.0038 | |
| GSE77861 | PDE4B | 5142 | 203708_at | 0.1701 | 0.4914 | |
| GSE97050 | PDE4B | 5142 | A_23_P74278 | 0.6115 | 0.1657 | |
| SRP007169 | PDE4B | 5142 | RNAseq | 1.1624 | 0.0373 | |
| SRP008496 | PDE4B | 5142 | RNAseq | 1.3904 | 0.0026 | |
| SRP064894 | PDE4B | 5142 | RNAseq | 0.5994 | 0.0985 | |
| SRP133303 | PDE4B | 5142 | RNAseq | 0.7664 | 0.0216 | |
| SRP159526 | PDE4B | 5142 | RNAseq | 1.4761 | 0.0086 | |
| SRP193095 | PDE4B | 5142 | RNAseq | 1.9293 | 0.0000 | |
| SRP219564 | PDE4B | 5142 | RNAseq | 1.0647 | 0.0035 | |
| TCGA | PDE4B | 5142 | RNAseq | -0.0180 | 0.9037 |
Upregulated datasets: 5; Downregulated datasets: 1.
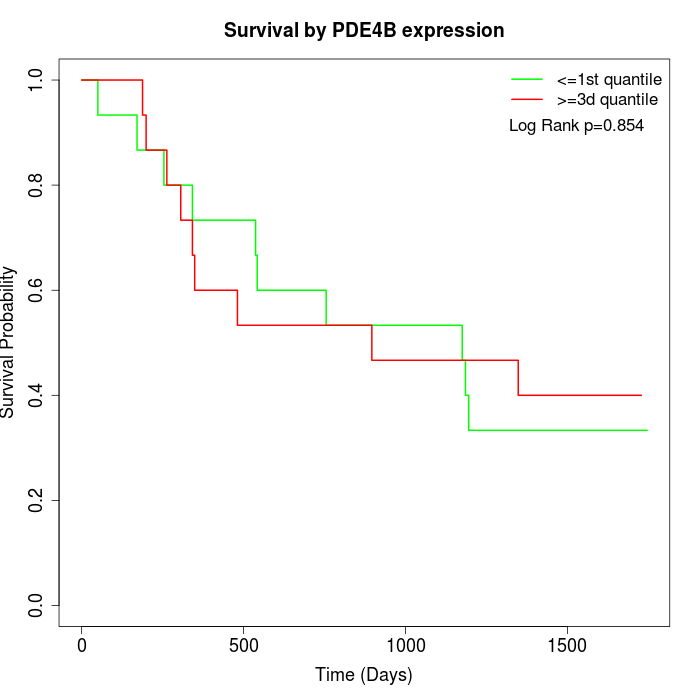
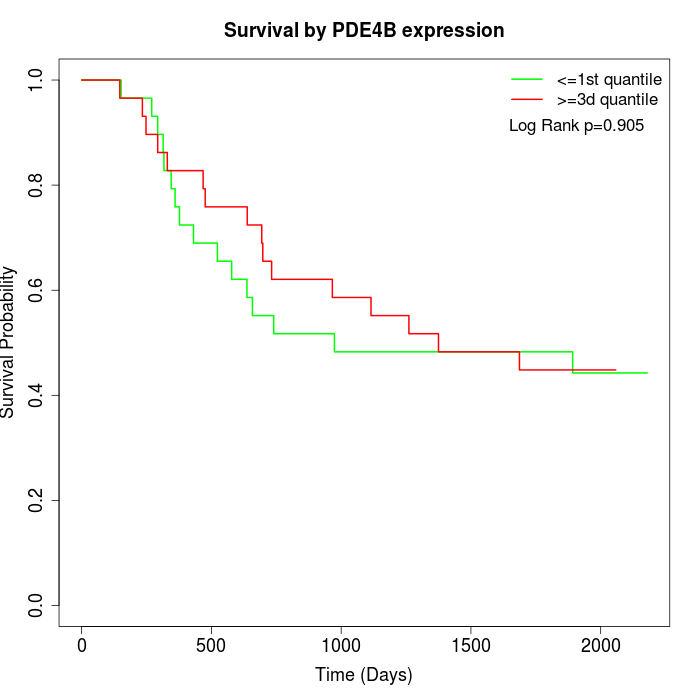
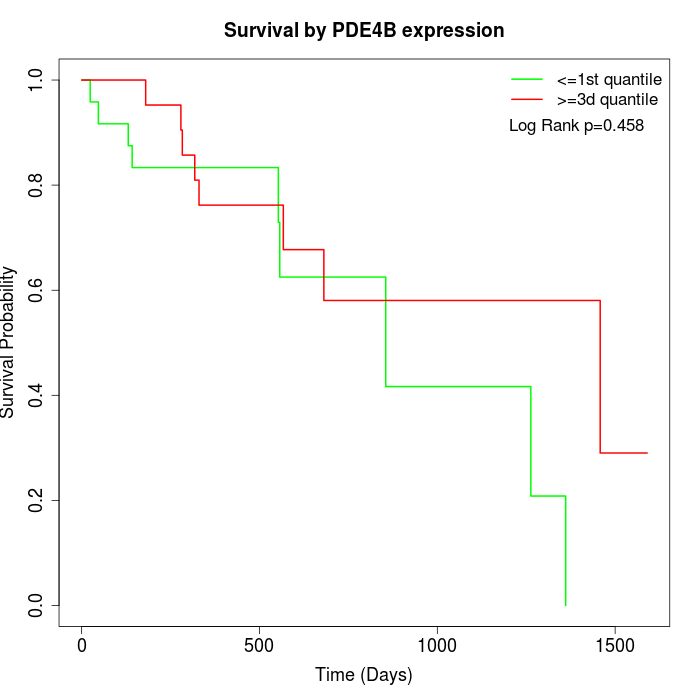
Survival by PDE4B expression:
Note: Click image to view full size file.
Copy number change of PDE4B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE4B | 5142 | 2 | 6 | 22 | |
| GSE20123 | PDE4B | 5142 | 2 | 5 | 23 | |
| GSE43470 | PDE4B | 5142 | 4 | 2 | 37 | |
| GSE46452 | PDE4B | 5142 | 0 | 1 | 58 | |
| GSE47630 | PDE4B | 5142 | 8 | 5 | 27 | |
| GSE54993 | PDE4B | 5142 | 0 | 0 | 70 | |
| GSE54994 | PDE4B | 5142 | 6 | 4 | 43 | |
| GSE60625 | PDE4B | 5142 | 0 | 0 | 11 | |
| GSE74703 | PDE4B | 5142 | 3 | 2 | 31 | |
| GSE74704 | PDE4B | 5142 | 1 | 2 | 17 | |
| TCGA | PDE4B | 5142 | 9 | 21 | 66 |
Total number of gains: 35; Total number of losses: 48; Total Number of normals: 405.
Somatic mutations of PDE4B:
Generating mutation plots.
Highly correlated genes for PDE4B:
Showing top 20/684 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE4B | GNGT2 | 0.812344 | 3 | 0 | 3 |
| PDE4B | GAPT | 0.747703 | 3 | 0 | 3 |
| PDE4B | PIGS | 0.742858 | 3 | 0 | 3 |
| PDE4B | SLAMF6 | 0.736332 | 3 | 0 | 3 |
| PDE4B | PARVG | 0.735614 | 3 | 0 | 3 |
| PDE4B | ZNF566 | 0.73126 | 3 | 0 | 3 |
| PDE4B | BTLA | 0.73094 | 3 | 0 | 3 |
| PDE4B | CLEC14A | 0.730643 | 4 | 0 | 4 |
| PDE4B | TNFSF13 | 0.715971 | 3 | 0 | 3 |
| PDE4B | CCL22 | 0.711467 | 3 | 0 | 3 |
| PDE4B | CCL11 | 0.70682 | 6 | 0 | 6 |
| PDE4B | CCR2 | 0.703616 | 4 | 0 | 3 |
| PDE4B | GBGT1 | 0.698175 | 4 | 0 | 3 |
| PDE4B | ZNF18 | 0.69757 | 3 | 0 | 3 |
| PDE4B | PREX1 | 0.694298 | 3 | 0 | 3 |
| PDE4B | CX3CR1 | 0.688309 | 3 | 0 | 3 |
| PDE4B | LRRK1 | 0.688158 | 4 | 0 | 4 |
| PDE4B | UNC5C | 0.687869 | 4 | 0 | 4 |
| PDE4B | ANPEP | 0.687699 | 4 | 0 | 3 |
| PDE4B | TRABD2A | 0.686969 | 3 | 0 | 3 |
For details and further investigation, click here