| Full name: regulator of G protein signaling 18 | Alias Symbol: RGS13 | ||
| Type: protein-coding gene | Cytoband: 1q31.2 | ||
| Entrez ID: 64407 | HGNC ID: HGNC:14261 | Ensembl Gene: ENSG00000150681 | OMIM ID: 607192 |
| Drug and gene relationship at DGIdb | |||
Expression of RGS18:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS18 | 64407 | 223809_at | 0.2901 | 0.8472 | |
| GSE26886 | RGS18 | 64407 | 223809_at | -0.2242 | 0.2980 | |
| GSE45670 | RGS18 | 64407 | 223809_at | -0.0476 | 0.9082 | |
| GSE53622 | RGS18 | 64407 | 47211 | -0.2102 | 0.3448 | |
| GSE53624 | RGS18 | 64407 | 47211 | -0.0959 | 0.6875 | |
| GSE63941 | RGS18 | 64407 | 223809_at | -2.6698 | 0.0000 | |
| GSE77861 | RGS18 | 64407 | 223809_at | -0.0145 | 0.8666 | |
| GSE97050 | RGS18 | 64407 | A_23_P302550 | 0.0254 | 0.9423 | |
| SRP007169 | RGS18 | 64407 | RNAseq | 0.9071 | 0.2629 | |
| SRP133303 | RGS18 | 64407 | RNAseq | 0.5030 | 0.1028 | |
| SRP219564 | RGS18 | 64407 | RNAseq | 0.4967 | 0.4047 | |
| TCGA | RGS18 | 64407 | RNAseq | 0.1688 | 0.4990 |
Upregulated datasets: 0; Downregulated datasets: 1.
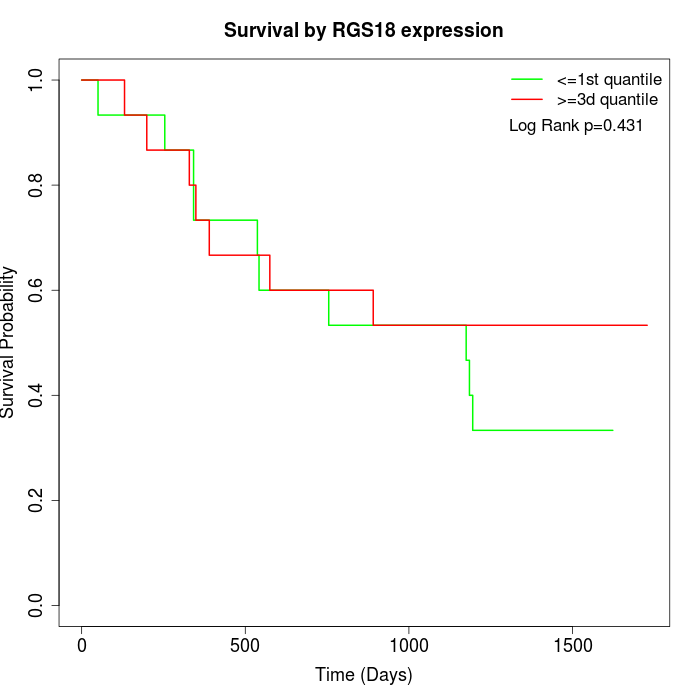
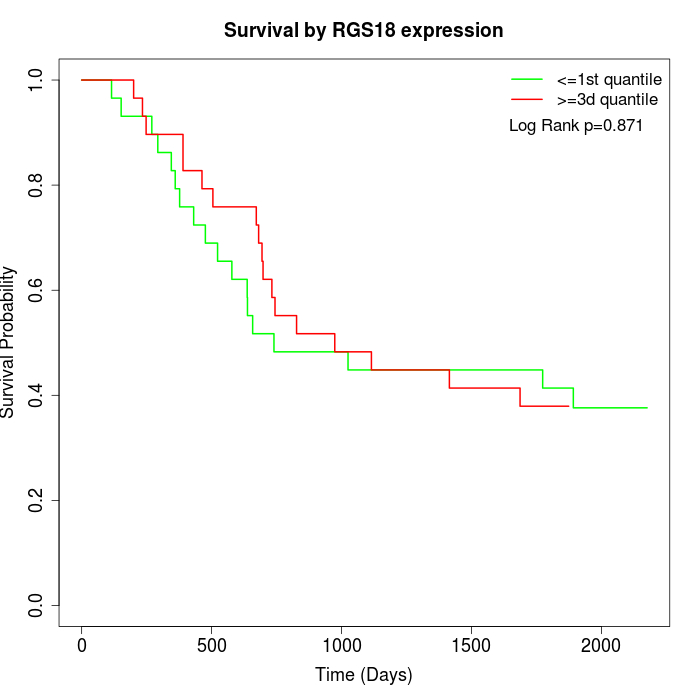
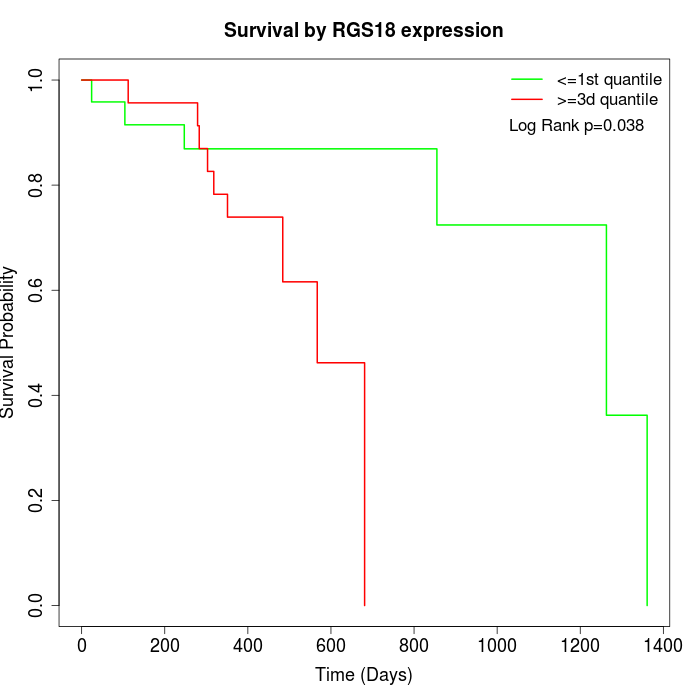
Survival by RGS18 expression:
Note: Click image to view full size file.
Copy number change of RGS18:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS18 | 64407 | 7 | 1 | 22 | |
| GSE20123 | RGS18 | 64407 | 7 | 1 | 22 | |
| GSE43470 | RGS18 | 64407 | 5 | 1 | 37 | |
| GSE46452 | RGS18 | 64407 | 3 | 1 | 55 | |
| GSE47630 | RGS18 | 64407 | 14 | 0 | 26 | |
| GSE54993 | RGS18 | 64407 | 0 | 6 | 64 | |
| GSE54994 | RGS18 | 64407 | 15 | 0 | 38 | |
| GSE60625 | RGS18 | 64407 | 0 | 0 | 11 | |
| GSE74703 | RGS18 | 64407 | 5 | 1 | 30 | |
| GSE74704 | RGS18 | 64407 | 2 | 1 | 17 | |
| TCGA | RGS18 | 64407 | 41 | 4 | 51 |
Total number of gains: 99; Total number of losses: 16; Total Number of normals: 373.
Somatic mutations of RGS18:
Generating mutation plots.
Highly correlated genes for RGS18:
Showing top 20/541 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS18 | C19orf38 | 0.84818 | 3 | 0 | 3 |
| RGS18 | GIMAP5 | 0.834368 | 3 | 0 | 3 |
| RGS18 | PRKD3 | 0.808131 | 3 | 0 | 3 |
| RGS18 | NPAT | 0.804721 | 3 | 0 | 3 |
| RGS18 | CCR2 | 0.783009 | 6 | 0 | 6 |
| RGS18 | ATP6V0D1 | 0.782004 | 3 | 0 | 3 |
| RGS18 | PHACTR1 | 0.777411 | 4 | 0 | 4 |
| RGS18 | IFT46 | 0.776028 | 3 | 0 | 3 |
| RGS18 | CLEC12A | 0.77313 | 4 | 0 | 4 |
| RGS18 | SAMM50 | 0.772823 | 3 | 0 | 3 |
| RGS18 | SNX20 | 0.767065 | 6 | 0 | 6 |
| RGS18 | CST7 | 0.764223 | 6 | 0 | 6 |
| RGS18 | FLI1 | 0.756006 | 7 | 0 | 6 |
| RGS18 | CYSLTR2 | 0.751431 | 4 | 0 | 4 |
| RGS18 | CD86 | 0.747463 | 3 | 0 | 3 |
| RGS18 | SIGLEC1 | 0.745798 | 3 | 0 | 3 |
| RGS18 | GIMAP6 | 0.745608 | 7 | 0 | 7 |
| RGS18 | C16orf54 | 0.745142 | 6 | 0 | 6 |
| RGS18 | GALM | 0.744091 | 3 | 0 | 3 |
| RGS18 | MTMR6 | 0.741756 | 3 | 0 | 3 |
For details and further investigation, click here