| Full name: cyclin dependent kinase inhibitor 2B | Alias Symbol: P15|MTS2|INK4B|TP15|CDK4I|p15INK4b | ||
| Type: protein-coding gene | Cytoband: 9p21.3 | ||
| Entrez ID: 1030 | HGNC ID: HGNC:1788 | Ensembl Gene: ENSG00000147883 | OMIM ID: 600431 |
| Drug and gene relationship at DGIdb | |||
CDKN2B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04110 | Cell cycle | |
| hsa04350 | TGF-beta signaling pathway | |
| hsa05166 | HTLV-I infection | |
| hsa05200 | Pathways in cancer | |
| hsa05222 | Small cell lung cancer |
Expression of CDKN2B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDKN2B | 1030 | 236313_at | -1.4159 | 0.3542 | |
| GSE20347 | CDKN2B | 1030 | 207530_s_at | -0.2893 | 0.0181 | |
| GSE23400 | CDKN2B | 1030 | 207530_s_at | -0.2825 | 0.0000 | |
| GSE26886 | CDKN2B | 1030 | 236313_at | -4.5108 | 0.0000 | |
| GSE29001 | CDKN2B | 1030 | 207530_s_at | -0.1334 | 0.3014 | |
| GSE38129 | CDKN2B | 1030 | 207530_s_at | -0.2845 | 0.0221 | |
| GSE45670 | CDKN2B | 1030 | 236313_at | -0.9857 | 0.3125 | |
| GSE53622 | CDKN2B | 1030 | 82442 | -1.6708 | 0.0000 | |
| GSE53624 | CDKN2B | 1030 | 82442 | -1.9325 | 0.0000 | |
| GSE63941 | CDKN2B | 1030 | 236313_at | -3.3941 | 0.0495 | |
| GSE77861 | CDKN2B | 1030 | 236313_at | -3.0522 | 0.0273 | |
| GSE97050 | CDKN2B | 1030 | A_23_P216812 | -0.6818 | 0.6244 | |
| SRP007169 | CDKN2B | 1030 | RNAseq | -1.6176 | 0.0040 | |
| SRP008496 | CDKN2B | 1030 | RNAseq | -1.4115 | 0.0022 | |
| SRP064894 | CDKN2B | 1030 | RNAseq | -0.4360 | 0.3199 | |
| SRP133303 | CDKN2B | 1030 | RNAseq | -1.1764 | 0.0560 | |
| SRP159526 | CDKN2B | 1030 | RNAseq | -3.0167 | 0.0000 | |
| SRP193095 | CDKN2B | 1030 | RNAseq | -3.0087 | 0.0000 | |
| SRP219564 | CDKN2B | 1030 | RNAseq | -2.3467 | 0.0032 | |
| TCGA | CDKN2B | 1030 | RNAseq | -0.0493 | 0.8481 |
Upregulated datasets: 0; Downregulated datasets: 10.
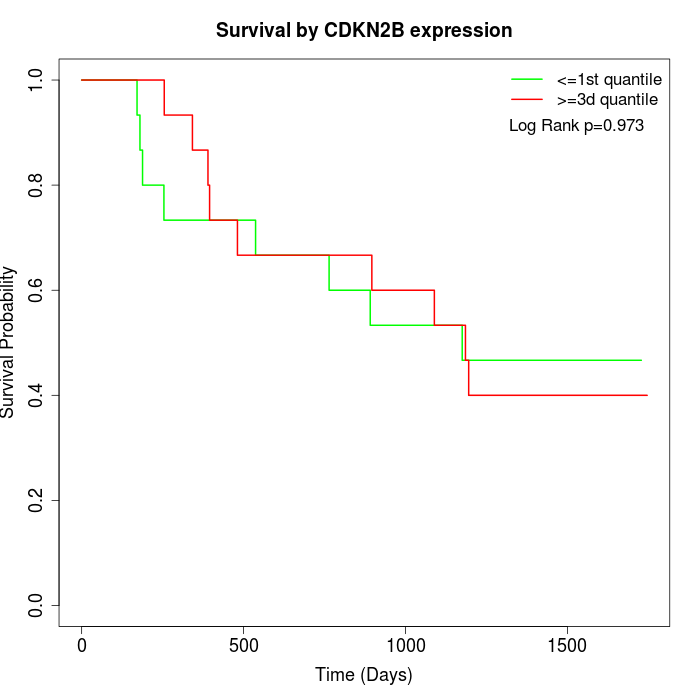
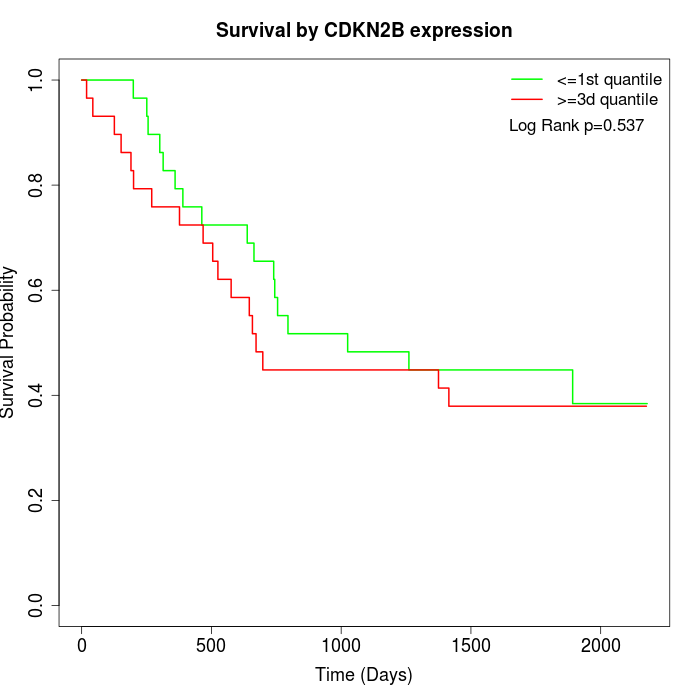
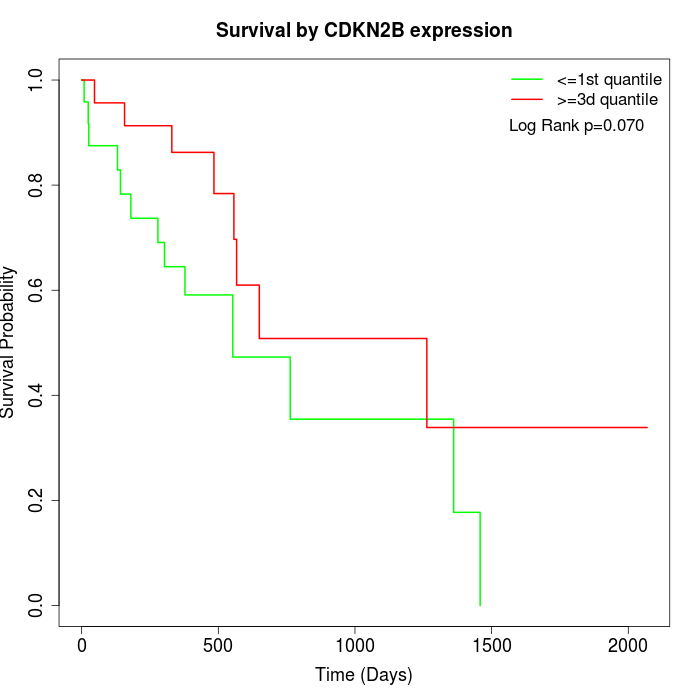
Survival by CDKN2B expression:
Note: Click image to view full size file.
Copy number change of CDKN2B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDKN2B | 1030 | 1 | 22 | 7 | |
| GSE20123 | CDKN2B | 1030 | 1 | 22 | 7 | |
| GSE43470 | CDKN2B | 1030 | 1 | 18 | 24 | |
| GSE46452 | CDKN2B | 1030 | 3 | 27 | 29 | |
| GSE47630 | CDKN2B | 1030 | 1 | 28 | 11 | |
| GSE54993 | CDKN2B | 1030 | 8 | 0 | 62 | |
| GSE54994 | CDKN2B | 1030 | 4 | 25 | 24 | |
| GSE60625 | CDKN2B | 1030 | 3 | 0 | 8 | |
| GSE74703 | CDKN2B | 1030 | 1 | 15 | 20 | |
| GSE74704 | CDKN2B | 1030 | 0 | 15 | 5 | |
| TCGA | CDKN2B | 1030 | 1 | 74 | 21 |
Total number of gains: 24; Total number of losses: 246; Total Number of normals: 218.
Somatic mutations of CDKN2B:
Generating mutation plots.
Highly correlated genes for CDKN2B:
Showing top 20/672 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDKN2B | CAPN14 | 0.764001 | 6 | 0 | 6 |
| CDKN2B | CYSRT1 | 0.743223 | 6 | 0 | 6 |
| CDKN2B | SH3PXD2A-AS1 | 0.731101 | 4 | 0 | 4 |
| CDKN2B | SMIM5 | 0.712321 | 6 | 0 | 6 |
| CDKN2B | TMEM154 | 0.711163 | 7 | 0 | 7 |
| CDKN2B | PCBP1-AS1 | 0.709632 | 3 | 0 | 3 |
| CDKN2B | ATG9B | 0.707075 | 6 | 0 | 6 |
| CDKN2B | C6orf132 | 0.702791 | 7 | 0 | 7 |
| CDKN2B | ARHGAP27 | 0.70253 | 6 | 0 | 6 |
| CDKN2B | SPNS2 | 0.698936 | 4 | 0 | 4 |
| CDKN2B | RAET1E | 0.69245 | 7 | 0 | 7 |
| CDKN2B | MORN3 | 0.682327 | 4 | 0 | 4 |
| CDKN2B | GRPEL2 | 0.680102 | 6 | 0 | 6 |
| CDKN2B | C4orf3 | 0.679834 | 6 | 0 | 6 |
| CDKN2B | SNORA68 | 0.679738 | 4 | 0 | 3 |
| CDKN2B | LIPH | 0.679605 | 7 | 0 | 6 |
| CDKN2B | PLEKHN1 | 0.677145 | 7 | 0 | 7 |
| CDKN2B | ATP13A4 | 0.67664 | 6 | 0 | 5 |
| CDKN2B | RAB10 | 0.676585 | 6 | 0 | 5 |
| CDKN2B | VSIG2 | 0.674126 | 7 | 0 | 6 |
For details and further investigation, click here