| Full name: cerebral dopamine neurotrophic factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10p13 | ||
| Entrez ID: 441549 | HGNC ID: HGNC:24913 | Ensembl Gene: ENSG00000185267 | OMIM ID: 611233 |
| Drug and gene relationship at DGIdb | |||
Expression of CDNF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDNF | 441549 | 1568696_at | -0.1102 | 0.6401 | |
| GSE26886 | CDNF | 441549 | 1568696_at | 0.1602 | 0.1021 | |
| GSE45670 | CDNF | 441549 | 1568696_at | -0.4586 | 0.0000 | |
| GSE53622 | CDNF | 441549 | 28008 | -0.9838 | 0.0000 | |
| GSE53624 | CDNF | 441549 | 28008 | -0.9725 | 0.0000 | |
| GSE63941 | CDNF | 441549 | 1568696_at | -0.2418 | 0.4825 | |
| GSE77861 | CDNF | 441549 | 1568696_at | -0.0927 | 0.2508 | |
| GSE97050 | CDNF | 441549 | A_24_P93309 | -0.4758 | 0.1279 | |
| SRP133303 | CDNF | 441549 | RNAseq | -0.6462 | 0.0982 | |
| SRP159526 | CDNF | 441549 | RNAseq | -0.8121 | 0.1042 | |
| SRP219564 | CDNF | 441549 | RNAseq | -0.2770 | 0.6805 | |
| TCGA | CDNF | 441549 | RNAseq | -0.9493 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 0.
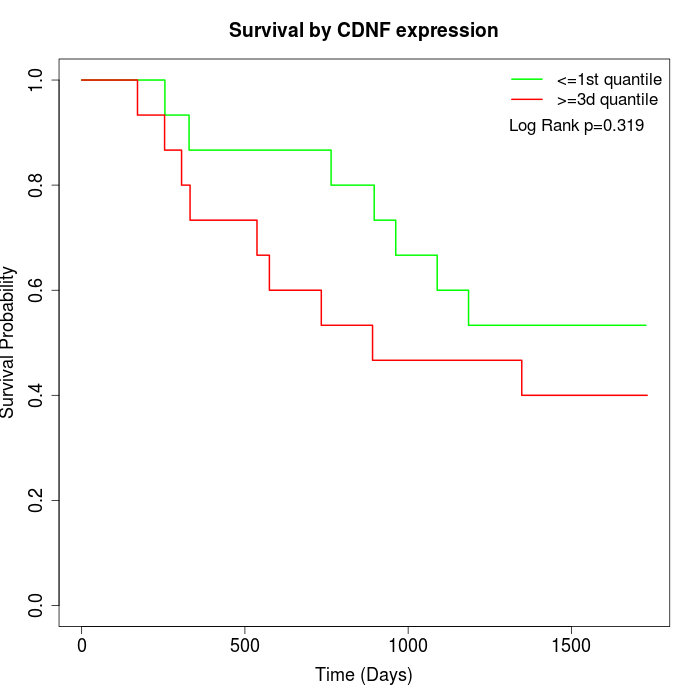
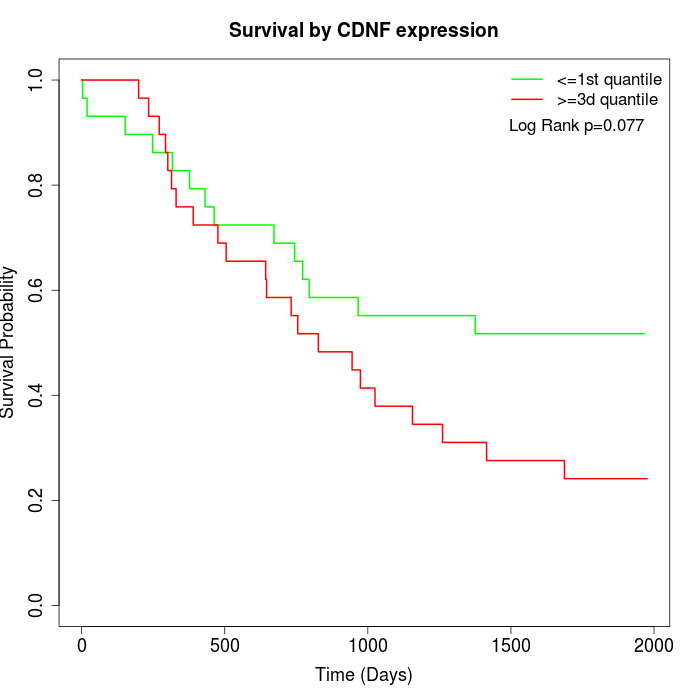
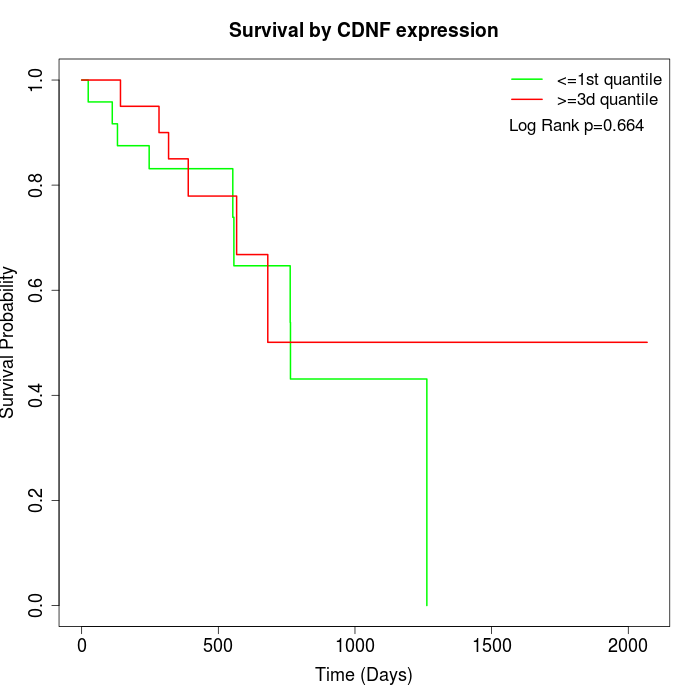
Survival by CDNF expression:
Note: Click image to view full size file.
Copy number change of CDNF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDNF | 441549 | 3 | 8 | 19 | |
| GSE20123 | CDNF | 441549 | 3 | 7 | 20 | |
| GSE43470 | CDNF | 441549 | 3 | 5 | 35 | |
| GSE46452 | CDNF | 441549 | 1 | 14 | 44 | |
| GSE47630 | CDNF | 441549 | 5 | 15 | 20 | |
| GSE54993 | CDNF | 441549 | 10 | 0 | 60 | |
| GSE54994 | CDNF | 441549 | 4 | 9 | 40 | |
| GSE60625 | CDNF | 441549 | 0 | 0 | 11 | |
| GSE74703 | CDNF | 441549 | 2 | 3 | 31 | |
| GSE74704 | CDNF | 441549 | 0 | 6 | 14 | |
| TCGA | CDNF | 441549 | 20 | 25 | 51 |
Total number of gains: 51; Total number of losses: 92; Total Number of normals: 345.
Somatic mutations of CDNF:
Generating mutation plots.
Highly correlated genes for CDNF:
Showing top 20/283 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDNF | FYTTD1 | 0.813461 | 3 | 0 | 3 |
| CDNF | DCUN1D4 | 0.776865 | 3 | 0 | 3 |
| CDNF | ABLIM2 | 0.770306 | 3 | 0 | 3 |
| CDNF | NPY1R | 0.754389 | 3 | 0 | 3 |
| CDNF | RNF111 | 0.747214 | 3 | 0 | 3 |
| CDNF | PRELP | 0.736868 | 3 | 0 | 3 |
| CDNF | UGP2 | 0.73 | 3 | 0 | 3 |
| CDNF | TMEM167B | 0.725299 | 3 | 0 | 3 |
| CDNF | GNG7 | 0.717656 | 4 | 0 | 4 |
| CDNF | RASL12 | 0.715915 | 3 | 0 | 3 |
| CDNF | MKRN2 | 0.706699 | 3 | 0 | 3 |
| CDNF | RAB18 | 0.706314 | 3 | 0 | 3 |
| CDNF | ANKRD6 | 0.703963 | 3 | 0 | 3 |
| CDNF | TDRD3 | 0.701143 | 4 | 0 | 4 |
| CDNF | VPS29 | 0.695718 | 4 | 0 | 4 |
| CDNF | NDUFA2 | 0.69028 | 4 | 0 | 3 |
| CDNF | DNAH1 | 0.690226 | 3 | 0 | 3 |
| CDNF | LYPLAL1 | 0.688506 | 4 | 0 | 3 |
| CDNF | AGGF1 | 0.686706 | 3 | 0 | 3 |
| CDNF | HBP1 | 0.685817 | 3 | 0 | 3 |
For details and further investigation, click here