| Full name: coiled-coil-helix-coiled-coil-helix domain containing 3 | Alias Symbol: FLJ20420|MINOS3|PPP1R22|Mic19|MICOS19 | ||
| Type: protein-coding gene | Cytoband: 7q32.3-q33 | ||
| Entrez ID: 54927 | HGNC ID: HGNC:21906 | Ensembl Gene: ENSG00000106554 | OMIM ID: 613748 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CHCHD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHCHD3 | 54927 | 217972_at | -0.2456 | 0.5820 | |
| GSE20347 | CHCHD3 | 54927 | 217972_at | 0.3610 | 0.0370 | |
| GSE23400 | CHCHD3 | 54927 | 217972_at | 0.3887 | 0.0000 | |
| GSE26886 | CHCHD3 | 54927 | 217972_at | -0.0069 | 0.9749 | |
| GSE29001 | CHCHD3 | 54927 | 217972_at | 0.3744 | 0.1893 | |
| GSE38129 | CHCHD3 | 54927 | 217972_at | 0.2634 | 0.0550 | |
| GSE45670 | CHCHD3 | 54927 | 217972_at | 0.0632 | 0.7789 | |
| GSE53622 | CHCHD3 | 54927 | 21796 | 0.3983 | 0.0000 | |
| GSE53624 | CHCHD3 | 54927 | 21796 | 0.4905 | 0.0000 | |
| GSE63941 | CHCHD3 | 54927 | 217972_at | -0.4430 | 0.2705 | |
| GSE77861 | CHCHD3 | 54927 | 217972_at | 0.3460 | 0.1220 | |
| GSE97050 | CHCHD3 | 54927 | A_23_P251927 | 0.0881 | 0.7898 | |
| SRP007169 | CHCHD3 | 54927 | RNAseq | 0.4716 | 0.1548 | |
| SRP008496 | CHCHD3 | 54927 | RNAseq | 0.7842 | 0.0001 | |
| SRP064894 | CHCHD3 | 54927 | RNAseq | 0.3181 | 0.0122 | |
| SRP133303 | CHCHD3 | 54927 | RNAseq | 0.4690 | 0.0000 | |
| SRP159526 | CHCHD3 | 54927 | RNAseq | 0.6626 | 0.0210 | |
| SRP193095 | CHCHD3 | 54927 | RNAseq | 0.2254 | 0.0778 | |
| SRP219564 | CHCHD3 | 54927 | RNAseq | -0.2143 | 0.6323 | |
| TCGA | CHCHD3 | 54927 | RNAseq | 0.0101 | 0.8664 |
Upregulated datasets: 0; Downregulated datasets: 0.
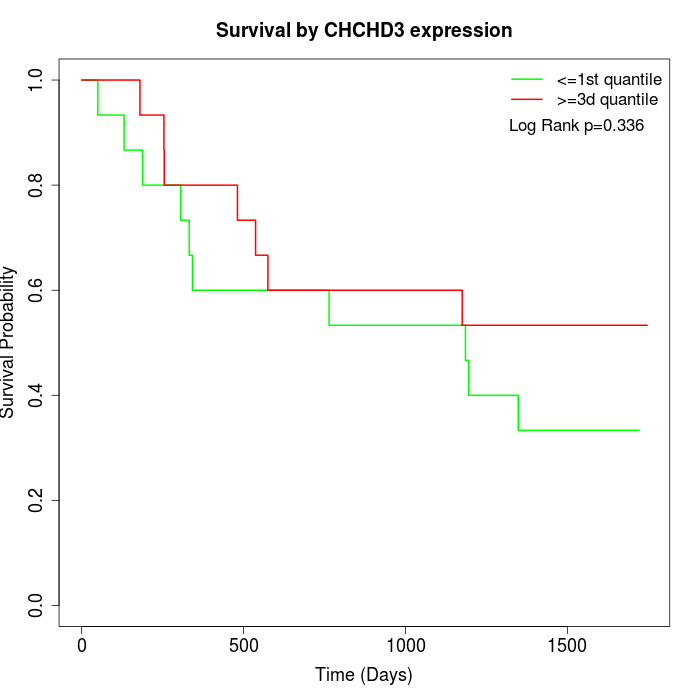
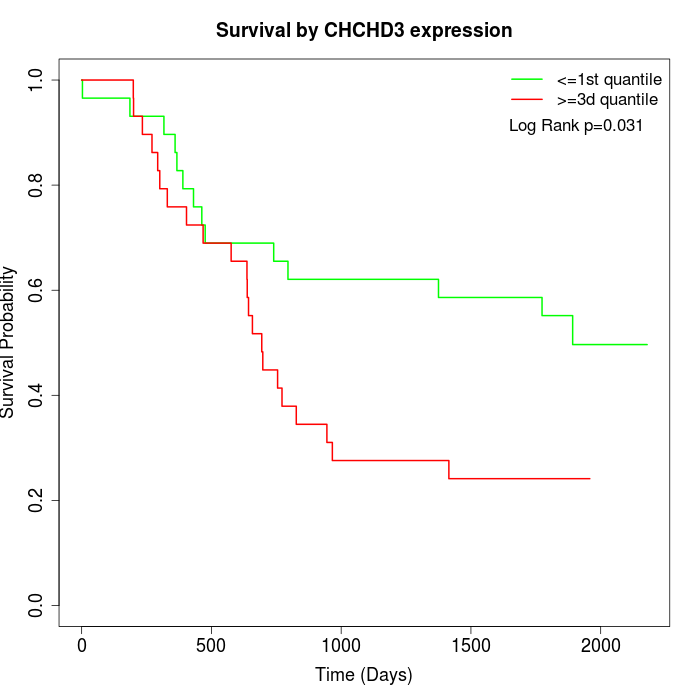
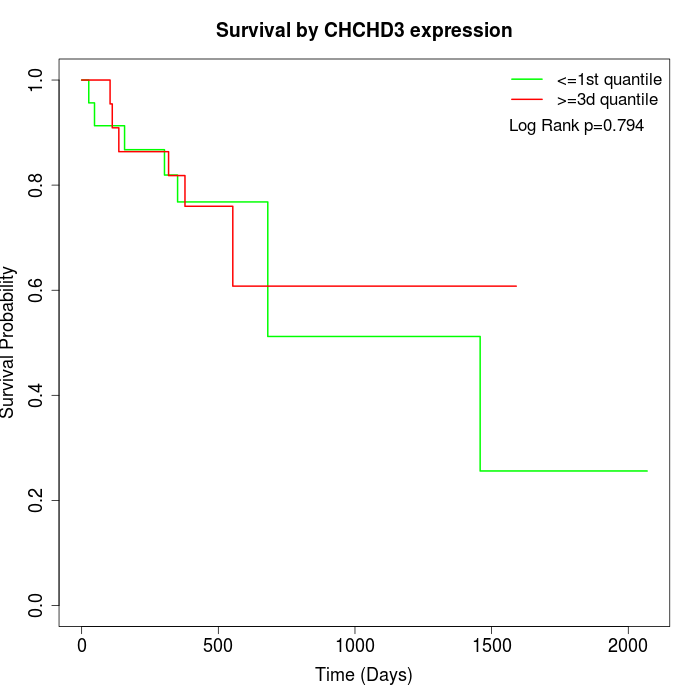
Survival by CHCHD3 expression:
Note: Click image to view full size file.
Copy number change of CHCHD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHCHD3 | 54927 | 4 | 2 | 24 | |
| GSE20123 | CHCHD3 | 54927 | 4 | 2 | 24 | |
| GSE43470 | CHCHD3 | 54927 | 2 | 3 | 38 | |
| GSE46452 | CHCHD3 | 54927 | 7 | 2 | 50 | |
| GSE47630 | CHCHD3 | 54927 | 7 | 4 | 29 | |
| GSE54993 | CHCHD3 | 54927 | 3 | 5 | 62 | |
| GSE54994 | CHCHD3 | 54927 | 6 | 7 | 40 | |
| GSE60625 | CHCHD3 | 54927 | 0 | 0 | 11 | |
| GSE74703 | CHCHD3 | 54927 | 2 | 3 | 31 | |
| GSE74704 | CHCHD3 | 54927 | 1 | 2 | 17 | |
| TCGA | CHCHD3 | 54927 | 28 | 22 | 46 |
Total number of gains: 64; Total number of losses: 52; Total Number of normals: 372.
Somatic mutations of CHCHD3:
Generating mutation plots.
Highly correlated genes for CHCHD3:
Showing top 20/672 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHCHD3 | CTPS2 | 0.787532 | 3 | 0 | 3 |
| CHCHD3 | ZNF304 | 0.714899 | 3 | 0 | 3 |
| CHCHD3 | PAPOLG | 0.71341 | 3 | 0 | 3 |
| CHCHD3 | MRPS5 | 0.710457 | 4 | 0 | 3 |
| CHCHD3 | MRPL2 | 0.70845 | 3 | 0 | 3 |
| CHCHD3 | RHEB | 0.70526 | 13 | 0 | 13 |
| CHCHD3 | COX10 | 0.705018 | 3 | 0 | 3 |
| CHCHD3 | MNS1 | 0.704224 | 3 | 0 | 3 |
| CHCHD3 | GPHN | 0.701979 | 3 | 0 | 3 |
| CHCHD3 | DERL2 | 0.694438 | 3 | 0 | 3 |
| CHCHD3 | JKAMP | 0.689419 | 3 | 0 | 3 |
| CHCHD3 | SPPL2A | 0.687559 | 3 | 0 | 3 |
| CHCHD3 | HIBADH | 0.686338 | 3 | 0 | 3 |
| CHCHD3 | ARTN | 0.685632 | 3 | 0 | 3 |
| CHCHD3 | ZNF800 | 0.685559 | 4 | 0 | 4 |
| CHCHD3 | PPIL3 | 0.680683 | 3 | 0 | 3 |
| CHCHD3 | ELAC2 | 0.678677 | 3 | 0 | 3 |
| CHCHD3 | RTF1 | 0.678021 | 4 | 0 | 3 |
| CHCHD3 | PNPLA8 | 0.674852 | 3 | 0 | 3 |
| CHCHD3 | LANCL2 | 0.674126 | 4 | 0 | 3 |
For details and further investigation, click here