| Full name: carbohydrate sulfotransferase 12 | Alias Symbol: C4S-2|C4ST2 | ||
| Type: protein-coding gene | Cytoband: 7p22.3 | ||
| Entrez ID: 55501 | HGNC ID: HGNC:17423 | Ensembl Gene: ENSG00000136213 | OMIM ID: 610129 |
| Drug and gene relationship at DGIdb | |||
Expression of CHST12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHST12 | 55501 | 218927_s_at | 0.6735 | 0.0510 | |
| GSE20347 | CHST12 | 55501 | 218927_s_at | 0.6242 | 0.0000 | |
| GSE23400 | CHST12 | 55501 | 218927_s_at | 0.4662 | 0.0000 | |
| GSE26886 | CHST12 | 55501 | 218927_s_at | 1.8853 | 0.0000 | |
| GSE29001 | CHST12 | 55501 | 218927_s_at | 0.4952 | 0.0030 | |
| GSE38129 | CHST12 | 55501 | 218927_s_at | 0.5365 | 0.0001 | |
| GSE45670 | CHST12 | 55501 | 218927_s_at | 0.1672 | 0.4371 | |
| GSE53622 | CHST12 | 55501 | 35960 | 0.6132 | 0.0000 | |
| GSE53624 | CHST12 | 55501 | 35960 | 0.8819 | 0.0000 | |
| GSE63941 | CHST12 | 55501 | 218927_s_at | -0.3981 | 0.4354 | |
| GSE77861 | CHST12 | 55501 | 218927_s_at | 0.3797 | 0.0155 | |
| GSE97050 | CHST12 | 55501 | A_33_P3373459 | 0.2401 | 0.2631 | |
| SRP007169 | CHST12 | 55501 | RNAseq | 0.5114 | 0.2112 | |
| SRP008496 | CHST12 | 55501 | RNAseq | 0.5038 | 0.0683 | |
| SRP064894 | CHST12 | 55501 | RNAseq | 0.7787 | 0.0013 | |
| SRP133303 | CHST12 | 55501 | RNAseq | -0.0817 | 0.2716 | |
| SRP159526 | CHST12 | 55501 | RNAseq | 0.5504 | 0.0490 | |
| SRP193095 | CHST12 | 55501 | RNAseq | 0.1744 | 0.1531 | |
| SRP219564 | CHST12 | 55501 | RNAseq | 0.3720 | 0.2159 | |
| TCGA | CHST12 | 55501 | RNAseq | 0.1324 | 0.0340 |
Upregulated datasets: 1; Downregulated datasets: 0.
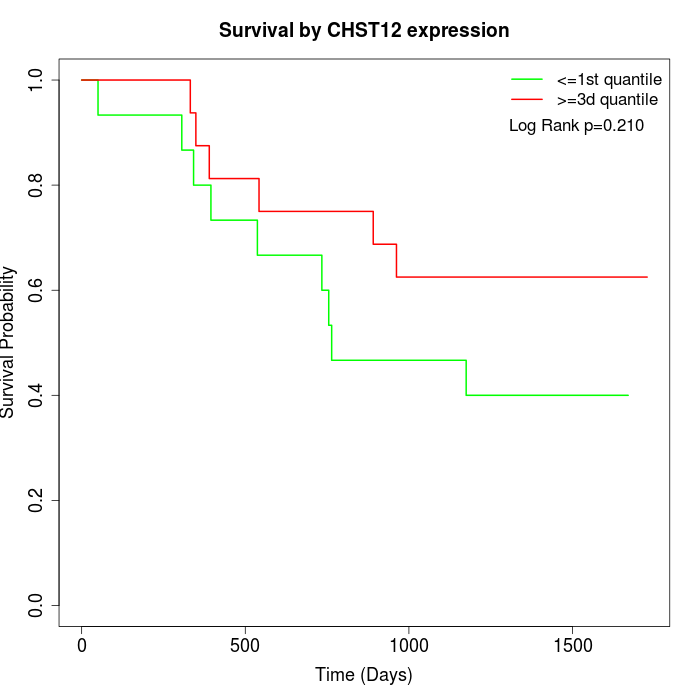
Survival by CHST12 expression:
Note: Click image to view full size file.
Copy number change of CHST12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHST12 | 55501 | 14 | 0 | 16 | |
| GSE20123 | CHST12 | 55501 | 14 | 0 | 16 | |
| GSE43470 | CHST12 | 55501 | 7 | 2 | 34 | |
| GSE46452 | CHST12 | 55501 | 13 | 1 | 45 | |
| GSE47630 | CHST12 | 55501 | 10 | 1 | 29 | |
| GSE54993 | CHST12 | 55501 | 0 | 7 | 63 | |
| GSE54994 | CHST12 | 55501 | 21 | 2 | 30 | |
| GSE60625 | CHST12 | 55501 | 0 | 0 | 11 | |
| GSE74703 | CHST12 | 55501 | 7 | 1 | 28 | |
| GSE74704 | CHST12 | 55501 | 9 | 0 | 11 | |
| TCGA | CHST12 | 55501 | 57 | 5 | 34 |
Total number of gains: 152; Total number of losses: 19; Total Number of normals: 317.
Somatic mutations of CHST12:
Generating mutation plots.
Highly correlated genes for CHST12:
Showing top 20/1711 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHST12 | RGS1 | 0.808584 | 3 | 0 | 3 |
| CHST12 | A4GALT | 0.804764 | 3 | 0 | 3 |
| CHST12 | CD44 | 0.763609 | 3 | 0 | 3 |
| CHST12 | PRR12 | 0.757639 | 3 | 0 | 3 |
| CHST12 | TP53I13 | 0.747877 | 6 | 0 | 6 |
| CHST12 | TMEM63B | 0.745943 | 3 | 0 | 3 |
| CHST12 | SLC46A1 | 0.742525 | 3 | 0 | 3 |
| CHST12 | PHF5A | 0.739177 | 4 | 0 | 3 |
| CHST12 | CHIC1 | 0.736678 | 3 | 0 | 3 |
| CHST12 | RAVER1 | 0.735133 | 3 | 0 | 3 |
| CHST12 | SCYL1 | 0.729706 | 4 | 0 | 4 |
| CHST12 | YWHAG | 0.727329 | 4 | 0 | 3 |
| CHST12 | KLHL13 | 0.727251 | 4 | 0 | 3 |
| CHST12 | KEAP1 | 0.726472 | 3 | 0 | 3 |
| CHST12 | RAB34 | 0.723396 | 3 | 0 | 3 |
| CHST12 | CDH24 | 0.723316 | 3 | 0 | 3 |
| CHST12 | CMTM4 | 0.72225 | 3 | 0 | 3 |
| CHST12 | ZNF496 | 0.721985 | 4 | 0 | 4 |
| CHST12 | PKIG | 0.720596 | 4 | 0 | 4 |
| CHST12 | BOLA3 | 0.719634 | 6 | 0 | 6 |
For details and further investigation, click here