| Full name: carbohydrate sulfotransferase 8 | Alias Symbol: GALNAC-4-ST1 | ||
| Type: protein-coding gene | Cytoband: 19q13.11 | ||
| Entrez ID: 64377 | HGNC ID: HGNC:15993 | Ensembl Gene: ENSG00000124302 | OMIM ID: 610190 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CHST8:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHST8 | 64377 | 221065_s_at | -0.0564 | 0.8206 | |
| GSE20347 | CHST8 | 64377 | 221065_s_at | -0.0555 | 0.4191 | |
| GSE23400 | CHST8 | 64377 | 221065_s_at | -0.1100 | 0.0019 | |
| GSE26886 | CHST8 | 64377 | 221065_s_at | 0.4354 | 0.0107 | |
| GSE29001 | CHST8 | 64377 | 221065_s_at | -0.2011 | 0.1821 | |
| GSE38129 | CHST8 | 64377 | 221065_s_at | -0.2257 | 0.0235 | |
| GSE45670 | CHST8 | 64377 | 221065_s_at | 0.0152 | 0.9095 | |
| GSE63941 | CHST8 | 64377 | 221065_s_at | 0.1823 | 0.4662 | |
| GSE77861 | CHST8 | 64377 | 221065_s_at | 0.0374 | 0.8221 | |
| TCGA | CHST8 | 64377 | RNAseq | -0.7680 | 0.1939 |
Upregulated datasets: 0; Downregulated datasets: 0.
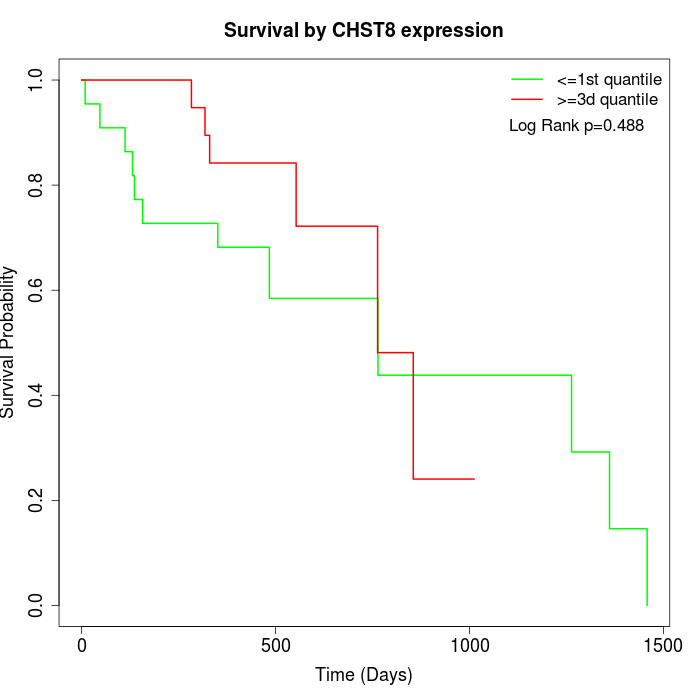
Survival by CHST8 expression:
Note: Click image to view full size file.
Copy number change of CHST8:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHST8 | 64377 | 7 | 6 | 17 | |
| GSE20123 | CHST8 | 64377 | 7 | 4 | 19 | |
| GSE43470 | CHST8 | 64377 | 5 | 6 | 32 | |
| GSE46452 | CHST8 | 64377 | 48 | 1 | 10 | |
| GSE47630 | CHST8 | 64377 | 8 | 6 | 26 | |
| GSE54993 | CHST8 | 64377 | 17 | 3 | 50 | |
| GSE54994 | CHST8 | 64377 | 8 | 9 | 36 | |
| GSE60625 | CHST8 | 64377 | 9 | 0 | 2 | |
| GSE74703 | CHST8 | 64377 | 5 | 4 | 27 | |
| GSE74704 | CHST8 | 64377 | 6 | 2 | 12 | |
| TCGA | CHST8 | 64377 | 18 | 12 | 66 |
Total number of gains: 138; Total number of losses: 53; Total Number of normals: 297.
Somatic mutations of CHST8:
Generating mutation plots.
Highly correlated genes for CHST8:
Showing top 20/831 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHST8 | MEIS3 | 0.745955 | 3 | 0 | 3 |
| CHST8 | LRRC46 | 0.737908 | 3 | 0 | 3 |
| CHST8 | RASL10B | 0.736495 | 3 | 0 | 3 |
| CHST8 | IGFALS | 0.729958 | 7 | 0 | 7 |
| CHST8 | EPO | 0.725251 | 5 | 0 | 5 |
| CHST8 | HOXB3 | 0.713343 | 3 | 0 | 3 |
| CHST8 | CDH23 | 0.707328 | 3 | 0 | 3 |
| CHST8 | PI16 | 0.703996 | 3 | 0 | 3 |
| CHST8 | LZTS1 | 0.702989 | 4 | 0 | 4 |
| CHST8 | DUSP9 | 0.700983 | 3 | 0 | 3 |
| CHST8 | LINC00598 | 0.700682 | 3 | 0 | 3 |
| CHST8 | TMEM145 | 0.696819 | 3 | 0 | 3 |
| CHST8 | MED14OS | 0.686709 | 3 | 0 | 3 |
| CHST8 | SPRED3 | 0.684368 | 3 | 0 | 3 |
| CHST8 | CLRN3 | 0.683355 | 3 | 0 | 3 |
| CHST8 | TMEM255B | 0.682044 | 3 | 0 | 3 |
| CHST8 | CHST13 | 0.680007 | 3 | 0 | 3 |
| CHST8 | HOXA11-AS | 0.678247 | 3 | 0 | 3 |
| CHST8 | OPCML | 0.677555 | 3 | 0 | 3 |
| CHST8 | FUT7 | 0.675767 | 6 | 0 | 5 |
For details and further investigation, click here