| Full name: insulin like growth factor binding protein acid labile subunit | Alias Symbol: ALS | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 3483 | HGNC ID: HGNC:5468 | Ensembl Gene: ENSG00000099769 | OMIM ID: 601489 |
| Drug and gene relationship at DGIdb | |||
Expression of IGFALS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IGFALS | 3483 | 215712_s_at | 0.0744 | 0.7120 | |
| GSE20347 | IGFALS | 3483 | 215712_s_at | -0.0313 | 0.7014 | |
| GSE23400 | IGFALS | 3483 | 215712_s_at | -0.0980 | 0.0118 | |
| GSE26886 | IGFALS | 3483 | 215712_s_at | 0.3756 | 0.0179 | |
| GSE29001 | IGFALS | 3483 | 215712_s_at | -0.1278 | 0.3832 | |
| GSE38129 | IGFALS | 3483 | 215712_s_at | -0.2304 | 0.0709 | |
| GSE45670 | IGFALS | 3483 | 215712_s_at | -0.0700 | 0.4445 | |
| GSE53622 | IGFALS | 3483 | 100758 | 0.5301 | 0.0000 | |
| GSE53624 | IGFALS | 3483 | 100758 | 0.5480 | 0.0000 | |
| GSE63941 | IGFALS | 3483 | 215712_s_at | 0.3807 | 0.0092 | |
| GSE77861 | IGFALS | 3483 | 215712_s_at | 0.0327 | 0.8814 | |
| GSE97050 | IGFALS | 3483 | A_23_P14892 | -0.1486 | 0.5672 | |
| TCGA | IGFALS | 3483 | RNAseq | -3.0774 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
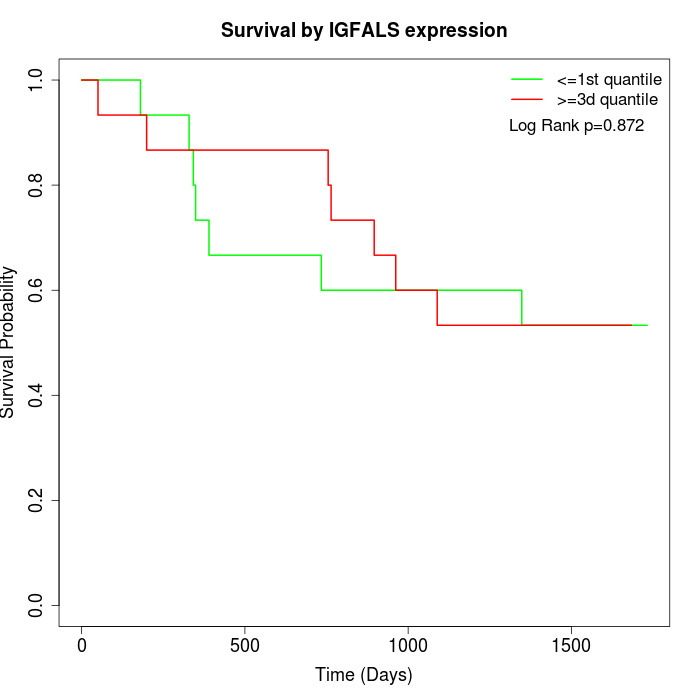
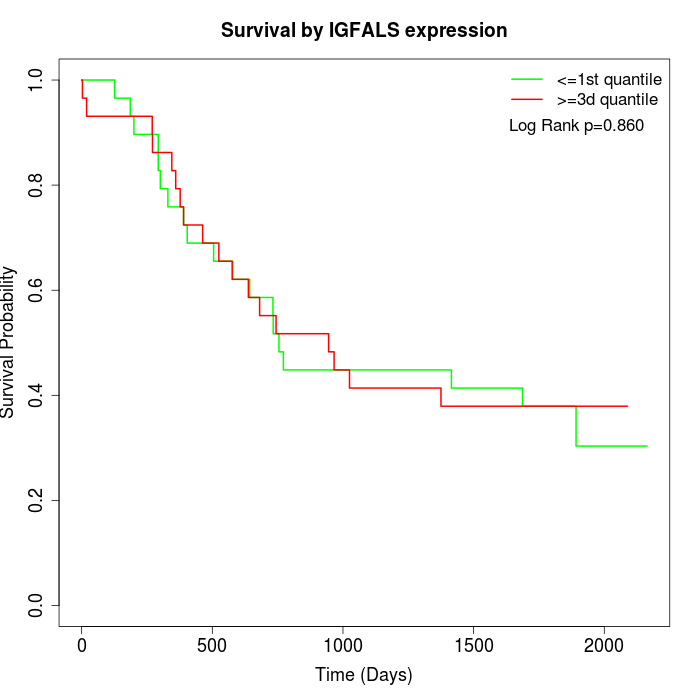
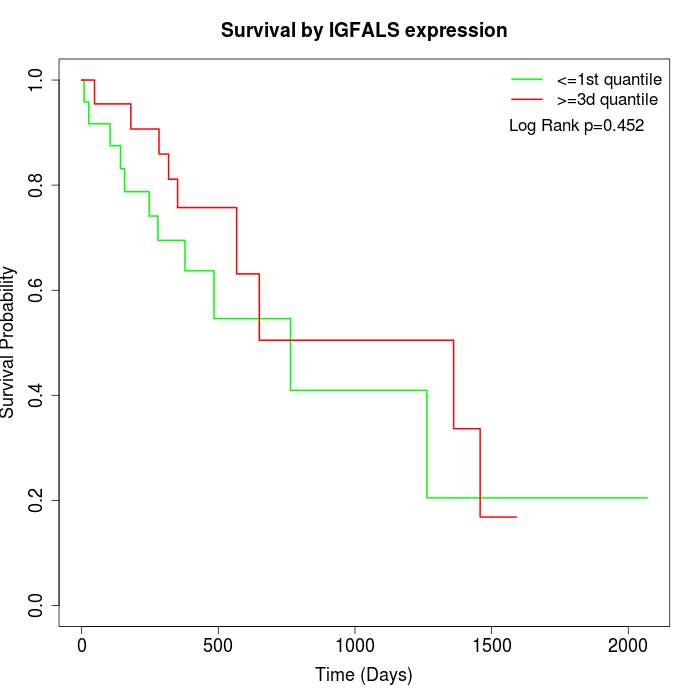
Survival by IGFALS expression:
Note: Click image to view full size file.
Copy number change of IGFALS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IGFALS | 3483 | 5 | 5 | 20 | |
| GSE20123 | IGFALS | 3483 | 5 | 4 | 21 | |
| GSE43470 | IGFALS | 3483 | 4 | 7 | 32 | |
| GSE46452 | IGFALS | 3483 | 38 | 1 | 20 | |
| GSE47630 | IGFALS | 3483 | 13 | 6 | 21 | |
| GSE54993 | IGFALS | 3483 | 3 | 5 | 62 | |
| GSE54994 | IGFALS | 3483 | 6 | 8 | 39 | |
| GSE60625 | IGFALS | 3483 | 4 | 0 | 7 | |
| GSE74703 | IGFALS | 3483 | 4 | 5 | 27 | |
| GSE74704 | IGFALS | 3483 | 3 | 2 | 15 | |
| TCGA | IGFALS | 3483 | 19 | 14 | 63 |
Total number of gains: 104; Total number of losses: 57; Total Number of normals: 327.
Somatic mutations of IGFALS:
Generating mutation plots.
Highly correlated genes for IGFALS:
Showing top 20/858 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IGFALS | PPP1R12C | 0.780635 | 3 | 0 | 3 |
| IGFALS | SP8 | 0.767468 | 3 | 0 | 3 |
| IGFALS | SBNO2 | 0.758957 | 3 | 0 | 3 |
| IGFALS | SCUBE1 | 0.754828 | 5 | 0 | 5 |
| IGFALS | BSND | 0.7508 | 3 | 0 | 3 |
| IGFALS | LENG1 | 0.741276 | 3 | 0 | 3 |
| IGFALS | HOXD9 | 0.74057 | 4 | 0 | 3 |
| IGFALS | FAM181B | 0.733316 | 3 | 0 | 3 |
| IGFALS | CHST8 | 0.729958 | 7 | 0 | 7 |
| IGFALS | ARMC5 | 0.719733 | 4 | 0 | 4 |
| IGFALS | LRFN2 | 0.719554 | 3 | 0 | 3 |
| IGFALS | MAG | 0.714415 | 7 | 0 | 7 |
| IGFALS | GPHB5 | 0.714063 | 3 | 0 | 3 |
| IGFALS | TEX22 | 0.713393 | 3 | 0 | 3 |
| IGFALS | RIMS4 | 0.708207 | 5 | 0 | 4 |
| IGFALS | SHARPIN | 0.707083 | 4 | 0 | 4 |
| IGFALS | REG3A | 0.705888 | 6 | 0 | 5 |
| IGFALS | HAO1 | 0.702042 | 3 | 0 | 3 |
| IGFALS | MRGPRG | 0.701887 | 3 | 0 | 3 |
| IGFALS | CDH23 | 0.700507 | 4 | 0 | 4 |
For details and further investigation, click here