| Full name: Charcot-Leyden crystal galectin | Alias Symbol: LGALS10|MGC149659|Gal-10 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 1178 | HGNC ID: HGNC:2014 | Ensembl Gene: ENSG00000105205 | OMIM ID: 153310 |
| Drug and gene relationship at DGIdb | |||
Expression of CLC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLC | 1178 | 206207_at | -0.0157 | 0.9945 | |
| GSE20347 | CLC | 1178 | 206207_at | 0.0534 | 0.5006 | |
| GSE23400 | CLC | 1178 | 206207_at | 0.0669 | 0.5047 | |
| GSE26886 | CLC | 1178 | 206207_at | -0.0295 | 0.8152 | |
| GSE29001 | CLC | 1178 | 206207_at | -0.1125 | 0.5852 | |
| GSE38129 | CLC | 1178 | 206207_at | 0.3301 | 0.0373 | |
| GSE45670 | CLC | 1178 | 206207_at | 0.2395 | 0.1452 | |
| GSE53622 | CLC | 1178 | 92104 | 0.9387 | 0.0782 | |
| GSE53624 | CLC | 1178 | 92104 | 0.4209 | 0.4204 | |
| GSE63941 | CLC | 1178 | 206207_at | 0.1924 | 0.7723 | |
| GSE77861 | CLC | 1178 | 206207_at | -0.0176 | 0.8752 | |
| GSE97050 | CLC | 1178 | A_23_P101683 | 0.5654 | 0.3770 | |
| TCGA | CLC | 1178 | RNAseq | -0.5056 | 0.6720 |
Upregulated datasets: 0; Downregulated datasets: 0.
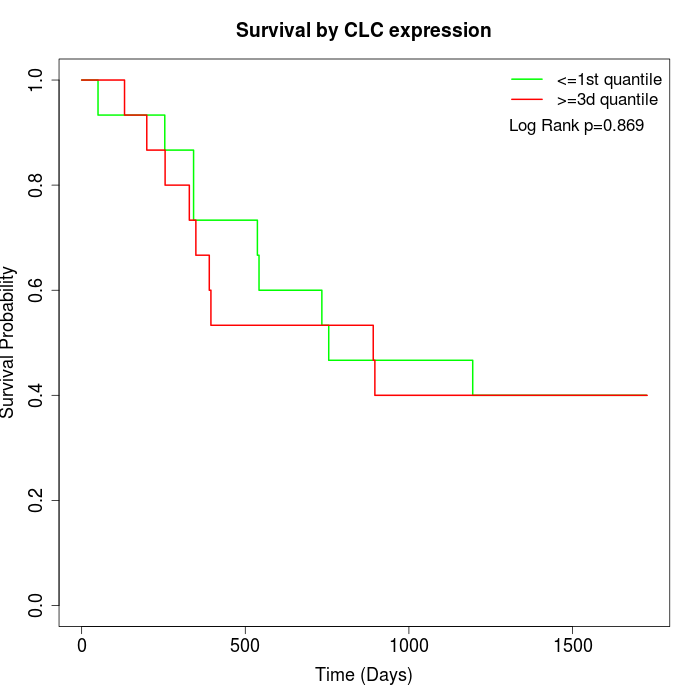
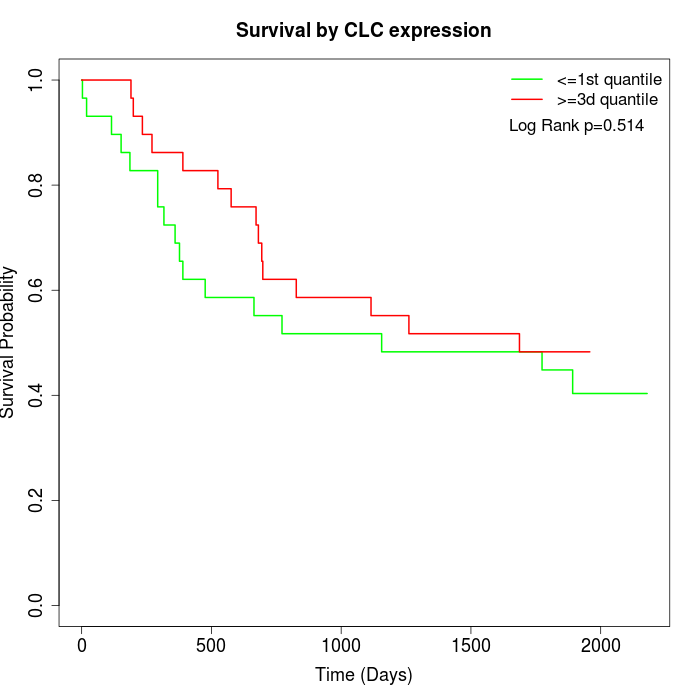
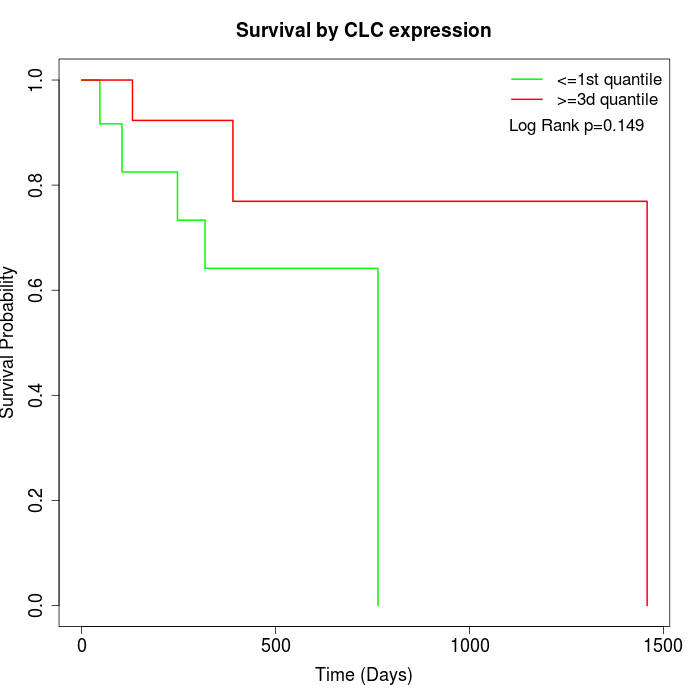
Survival by CLC expression:
Note: Click image to view full size file.
Copy number change of CLC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLC | 1178 | 7 | 4 | 19 | |
| GSE20123 | CLC | 1178 | 7 | 3 | 20 | |
| GSE43470 | CLC | 1178 | 3 | 11 | 29 | |
| GSE46452 | CLC | 1178 | 48 | 1 | 10 | |
| GSE47630 | CLC | 1178 | 9 | 5 | 26 | |
| GSE54993 | CLC | 1178 | 17 | 3 | 50 | |
| GSE54994 | CLC | 1178 | 7 | 9 | 37 | |
| GSE60625 | CLC | 1178 | 9 | 0 | 2 | |
| GSE74703 | CLC | 1178 | 3 | 7 | 26 | |
| GSE74704 | CLC | 1178 | 7 | 1 | 12 | |
| TCGA | CLC | 1178 | 14 | 16 | 66 |
Total number of gains: 131; Total number of losses: 60; Total Number of normals: 297.
Somatic mutations of CLC:
Generating mutation plots.
Highly correlated genes for CLC:
Showing top 20/321 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLC | MTAP | 0.811113 | 3 | 0 | 3 |
| CLC | TAF6L | 0.789731 | 3 | 0 | 3 |
| CLC | GDF5 | 0.761746 | 3 | 0 | 3 |
| CLC | RNASE3 | 0.749139 | 3 | 0 | 3 |
| CLC | GPR34 | 0.746063 | 4 | 0 | 4 |
| CLC | KCNK2 | 0.736699 | 3 | 0 | 3 |
| CLC | CPA3 | 0.73625 | 3 | 0 | 3 |
| CLC | STAT2 | 0.72539 | 3 | 0 | 3 |
| CLC | ZNF324B | 0.723229 | 3 | 0 | 3 |
| CLC | KCNQ3 | 0.7211 | 3 | 0 | 3 |
| CLC | MYT1 | 0.713041 | 3 | 0 | 3 |
| CLC | MAGI1 | 0.713005 | 3 | 0 | 3 |
| CLC | OR12D2 | 0.710905 | 3 | 0 | 3 |
| CLC | BTN1A1 | 0.704608 | 3 | 0 | 3 |
| CLC | MAP1LC3C | 0.702972 | 3 | 0 | 3 |
| CLC | KCNJ14 | 0.702516 | 3 | 0 | 3 |
| CLC | CCL8 | 0.702329 | 3 | 0 | 3 |
| CLC | OR7E24 | 0.701888 | 3 | 0 | 3 |
| CLC | BAGE | 0.699413 | 4 | 0 | 4 |
| CLC | ITGB3 | 0.69398 | 4 | 0 | 4 |
For details and further investigation, click here