| Full name: calmegin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q31.1 | ||
| Entrez ID: 1047 | HGNC ID: HGNC:2060 | Ensembl Gene: ENSG00000153132 | OMIM ID: 601858 |
| Drug and gene relationship at DGIdb | |||
Expression of CLGN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLGN | 1047 | 205830_at | -1.0327 | 0.2679 | |
| GSE20347 | CLGN | 1047 | 205830_at | -0.1803 | 0.4820 | |
| GSE23400 | CLGN | 1047 | 205830_at | -0.0600 | 0.1147 | |
| GSE26886 | CLGN | 1047 | 205830_at | -0.2825 | 0.6091 | |
| GSE29001 | CLGN | 1047 | 205830_at | -0.4059 | 0.5643 | |
| GSE38129 | CLGN | 1047 | 205830_at | -0.1221 | 0.6892 | |
| GSE45670 | CLGN | 1047 | 205830_at | -1.9587 | 0.0007 | |
| GSE53622 | CLGN | 1047 | 9755 | -1.0347 | 0.0000 | |
| GSE53624 | CLGN | 1047 | 9755 | -1.0029 | 0.0000 | |
| GSE63941 | CLGN | 1047 | 205830_at | -2.0079 | 0.2323 | |
| GSE77861 | CLGN | 1047 | 205830_at | 0.1134 | 0.5880 | |
| GSE97050 | CLGN | 1047 | A_23_P18684 | -0.2422 | 0.3727 | |
| SRP007169 | CLGN | 1047 | RNAseq | -3.5063 | 0.0004 | |
| SRP064894 | CLGN | 1047 | RNAseq | -2.2950 | 0.0000 | |
| SRP133303 | CLGN | 1047 | RNAseq | 0.2391 | 0.6479 | |
| SRP159526 | CLGN | 1047 | RNAseq | 0.2740 | 0.6472 | |
| SRP219564 | CLGN | 1047 | RNAseq | -1.7076 | 0.0101 | |
| TCGA | CLGN | 1047 | RNAseq | -0.2912 | 0.5294 |
Upregulated datasets: 0; Downregulated datasets: 6.
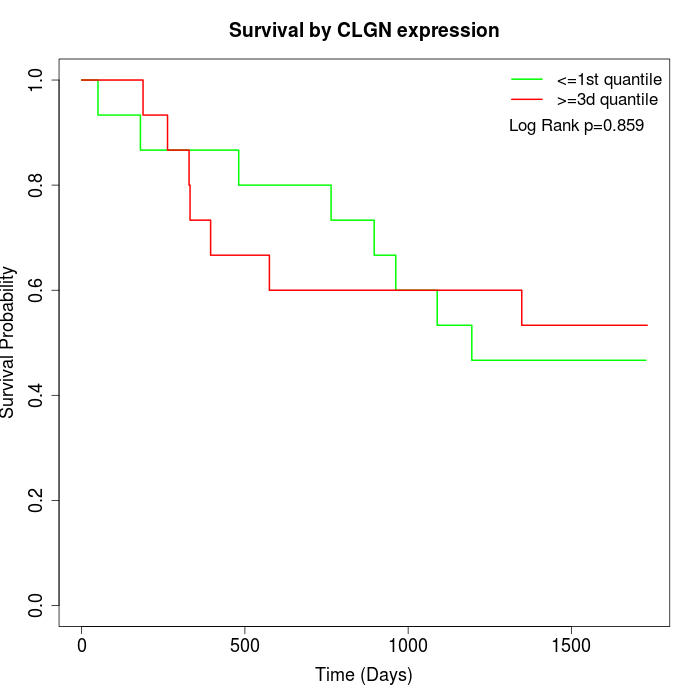
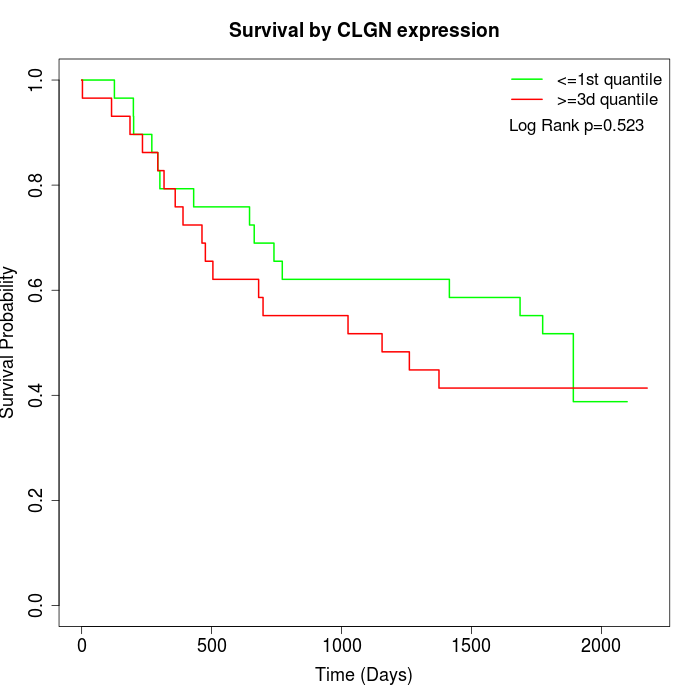
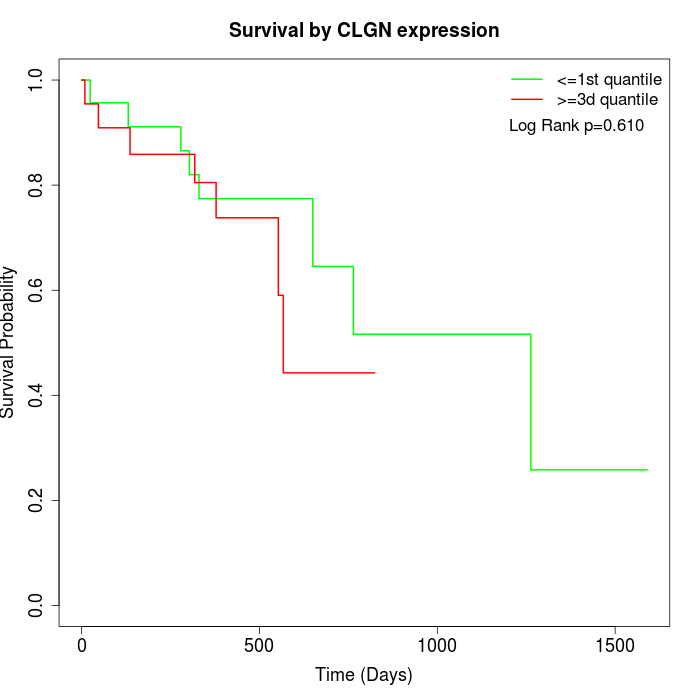
Survival by CLGN expression:
Note: Click image to view full size file.
Copy number change of CLGN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLGN | 1047 | 0 | 12 | 18 | |
| GSE20123 | CLGN | 1047 | 0 | 12 | 18 | |
| GSE43470 | CLGN | 1047 | 0 | 13 | 30 | |
| GSE46452 | CLGN | 1047 | 1 | 36 | 22 | |
| GSE47630 | CLGN | 1047 | 0 | 22 | 18 | |
| GSE54993 | CLGN | 1047 | 10 | 0 | 60 | |
| GSE54994 | CLGN | 1047 | 2 | 11 | 40 | |
| GSE60625 | CLGN | 1047 | 0 | 1 | 10 | |
| GSE74703 | CLGN | 1047 | 0 | 11 | 25 | |
| GSE74704 | CLGN | 1047 | 0 | 6 | 14 | |
| TCGA | CLGN | 1047 | 12 | 31 | 53 |
Total number of gains: 25; Total number of losses: 155; Total Number of normals: 308.
Somatic mutations of CLGN:
Generating mutation plots.
Highly correlated genes for CLGN:
Showing top 20/98 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLGN | ANKDD1A | 0.690676 | 3 | 0 | 3 |
| CLGN | VAMP4 | 0.66413 | 4 | 0 | 3 |
| CLGN | ZNF428 | 0.654191 | 4 | 0 | 4 |
| CLGN | TNNC2 | 0.622373 | 3 | 0 | 3 |
| CLGN | MAATS1 | 0.617222 | 4 | 0 | 3 |
| CLGN | ZNF568 | 0.614734 | 3 | 0 | 3 |
| CLGN | ABCB1 | 0.607852 | 5 | 0 | 4 |
| CLGN | TRPS1 | 0.60057 | 7 | 0 | 7 |
| CLGN | PEBP1 | 0.598446 | 4 | 0 | 3 |
| CLGN | HMGCLL1 | 0.592444 | 6 | 0 | 3 |
| CLGN | CRTAC1 | 0.590918 | 6 | 0 | 4 |
| CLGN | CALCOCO1 | 0.590835 | 5 | 0 | 3 |
| CLGN | MTERF2 | 0.590548 | 4 | 0 | 3 |
| CLGN | NTN1 | 0.590072 | 4 | 0 | 3 |
| CLGN | DDAH2 | 0.587514 | 5 | 0 | 4 |
| CLGN | ISCU | 0.587158 | 5 | 0 | 4 |
| CLGN | TRIM2 | 0.585856 | 7 | 0 | 5 |
| CLGN | ZRSR2 | 0.584539 | 3 | 0 | 3 |
| CLGN | C16orf89 | 0.582175 | 4 | 0 | 3 |
| CLGN | FAM189A2 | 0.581357 | 6 | 0 | 3 |
For details and further investigation, click here