| Full name: tripartite motif containing 2 | Alias Symbol: KIAA0517|RNF86|CMT2R | ||
| Type: protein-coding gene | Cytoband: 4q31.3 | ||
| Entrez ID: 23321 | HGNC ID: HGNC:15974 | Ensembl Gene: ENSG00000109654 | OMIM ID: 614141 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM2 | 23321 | 202341_s_at | -0.5430 | 0.6341 | |
| GSE20347 | TRIM2 | 23321 | 202342_s_at | -0.3559 | 0.4404 | |
| GSE23400 | TRIM2 | 23321 | 202341_s_at | -0.2488 | 0.0729 | |
| GSE26886 | TRIM2 | 23321 | 202341_s_at | -0.4106 | 0.4277 | |
| GSE29001 | TRIM2 | 23321 | 202341_s_at | -0.9090 | 0.2207 | |
| GSE38129 | TRIM2 | 23321 | 202342_s_at | -0.3800 | 0.2536 | |
| GSE45670 | TRIM2 | 23321 | 202341_s_at | -0.3633 | 0.2982 | |
| GSE53622 | TRIM2 | 23321 | 53033 | -0.5905 | 0.0000 | |
| GSE53624 | TRIM2 | 23321 | 53033 | -0.8070 | 0.0000 | |
| GSE63941 | TRIM2 | 23321 | 202342_s_at | -1.8828 | 0.0272 | |
| GSE77861 | TRIM2 | 23321 | 202342_s_at | -0.0240 | 0.9703 | |
| GSE97050 | TRIM2 | 23321 | A_24_P208909 | -0.4235 | 0.3154 | |
| SRP007169 | TRIM2 | 23321 | RNAseq | -1.0063 | 0.0178 | |
| SRP008496 | TRIM2 | 23321 | RNAseq | -0.8392 | 0.0219 | |
| SRP064894 | TRIM2 | 23321 | RNAseq | -1.2214 | 0.0020 | |
| SRP133303 | TRIM2 | 23321 | RNAseq | 0.5150 | 0.1238 | |
| SRP159526 | TRIM2 | 23321 | RNAseq | 0.5775 | 0.0964 | |
| SRP193095 | TRIM2 | 23321 | RNAseq | -0.2106 | 0.3776 | |
| SRP219564 | TRIM2 | 23321 | RNAseq | -1.2149 | 0.0737 | |
| TCGA | TRIM2 | 23321 | RNAseq | -0.3240 | 0.0022 |
Upregulated datasets: 0; Downregulated datasets: 3.
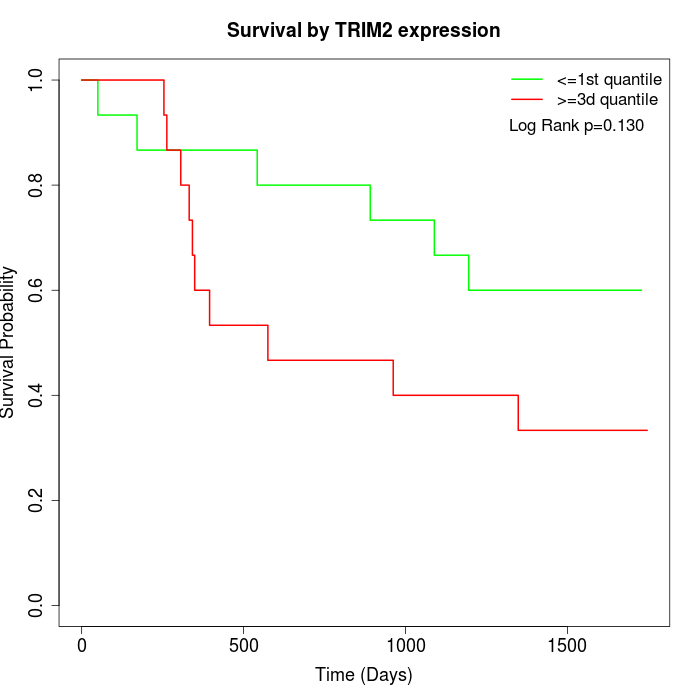
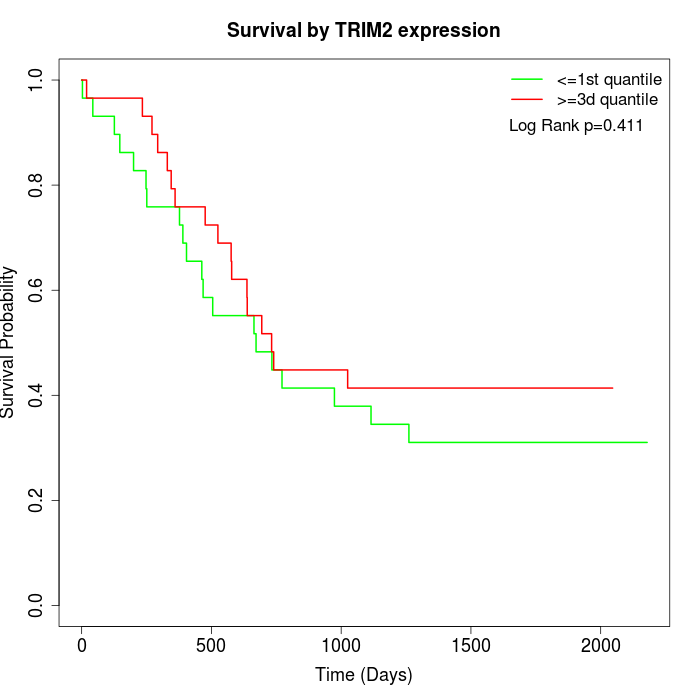
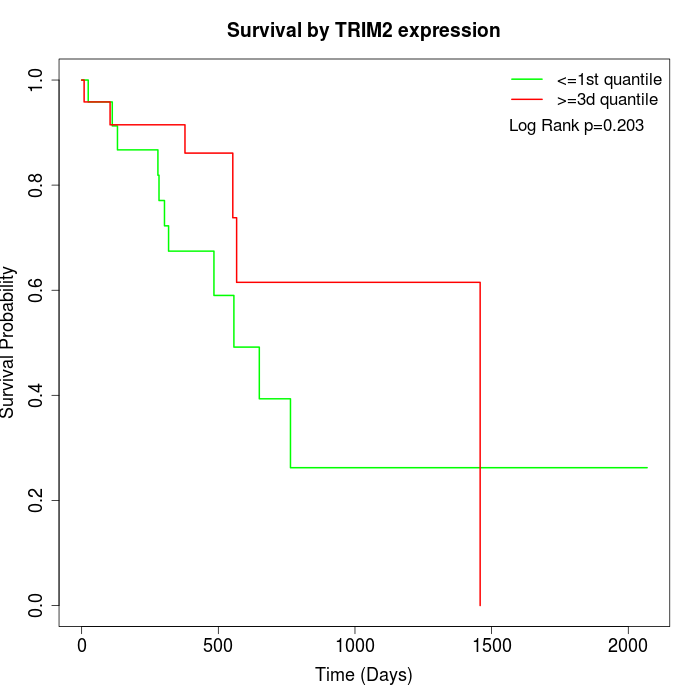
Survival by TRIM2 expression:
Note: Click image to view full size file.
Copy number change of TRIM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM2 | 23321 | 0 | 14 | 16 | |
| GSE20123 | TRIM2 | 23321 | 0 | 14 | 16 | |
| GSE43470 | TRIM2 | 23321 | 0 | 13 | 30 | |
| GSE46452 | TRIM2 | 23321 | 1 | 36 | 22 | |
| GSE47630 | TRIM2 | 23321 | 0 | 23 | 17 | |
| GSE54993 | TRIM2 | 23321 | 10 | 0 | 60 | |
| GSE54994 | TRIM2 | 23321 | 1 | 13 | 39 | |
| GSE60625 | TRIM2 | 23321 | 0 | 1 | 10 | |
| GSE74703 | TRIM2 | 23321 | 0 | 11 | 25 | |
| GSE74704 | TRIM2 | 23321 | 0 | 7 | 13 | |
| TCGA | TRIM2 | 23321 | 10 | 36 | 50 |
Total number of gains: 22; Total number of losses: 168; Total Number of normals: 298.
Somatic mutations of TRIM2:
Generating mutation plots.
Highly correlated genes for TRIM2:
Showing top 20/75 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM2 | BCDIN3D | 0.740688 | 3 | 0 | 3 |
| TRIM2 | CLHC1 | 0.698459 | 3 | 0 | 3 |
| TRIM2 | FHIT | 0.674768 | 3 | 0 | 3 |
| TRIM2 | ZNF682 | 0.67244 | 4 | 0 | 4 |
| TRIM2 | MID1 | 0.657983 | 4 | 0 | 3 |
| TRIM2 | GDF15 | 0.653421 | 3 | 0 | 3 |
| TRIM2 | TRDMT1 | 0.652656 | 3 | 0 | 3 |
| TRIM2 | JMJD8 | 0.642941 | 4 | 0 | 3 |
| TRIM2 | ZNF48 | 0.64233 | 3 | 0 | 3 |
| TRIM2 | PHKA2 | 0.641936 | 3 | 0 | 3 |
| TRIM2 | PRAMEF12 | 0.637146 | 3 | 0 | 3 |
| TRIM2 | PPCDC | 0.635042 | 3 | 0 | 3 |
| TRIM2 | ALDH1L1 | 0.626767 | 4 | 0 | 3 |
| TRIM2 | ACSS1 | 0.619919 | 4 | 0 | 3 |
| TRIM2 | NICN1 | 0.611475 | 4 | 0 | 3 |
| TRIM2 | C16orf89 | 0.602403 | 4 | 0 | 3 |
| TRIM2 | ZBTB6 | 0.600375 | 3 | 0 | 3 |
| TRIM2 | CCL17 | 0.594763 | 4 | 0 | 3 |
| TRIM2 | GRB7 | 0.594289 | 3 | 0 | 3 |
| TRIM2 | RNF20 | 0.591881 | 3 | 0 | 3 |
For details and further investigation, click here