| Full name: clathrin light chain A | Alias Symbol: Lca | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 1211 | HGNC ID: HGNC:2090 | Ensembl Gene: ENSG00000122705 | OMIM ID: 118960 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CLTA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05016 | Huntington's disease |
Expression of CLTA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLTA | 1211 | 200960_x_at | -0.2135 | 0.5350 | |
| GSE20347 | CLTA | 1211 | 200960_x_at | -0.2724 | 0.0966 | |
| GSE23400 | CLTA | 1211 | 200960_x_at | 0.0011 | 0.9884 | |
| GSE26886 | CLTA | 1211 | 216295_s_at | -0.0157 | 0.9270 | |
| GSE29001 | CLTA | 1211 | 200960_x_at | -0.3260 | 0.1142 | |
| GSE38129 | CLTA | 1211 | 200960_x_at | -0.0683 | 0.6169 | |
| GSE45670 | CLTA | 1211 | 200960_x_at | 0.0190 | 0.9074 | |
| GSE53622 | CLTA | 1211 | 59624 | -0.2710 | 0.0001 | |
| GSE53624 | CLTA | 1211 | 59624 | -0.0377 | 0.6919 | |
| GSE63941 | CLTA | 1211 | 200960_x_at | -0.3458 | 0.3572 | |
| GSE77861 | CLTA | 1211 | 200960_x_at | -0.3031 | 0.1842 | |
| GSE97050 | CLTA | 1211 | A_23_P219144 | -0.1016 | 0.7769 | |
| SRP007169 | CLTA | 1211 | RNAseq | -0.8055 | 0.0804 | |
| SRP008496 | CLTA | 1211 | RNAseq | -0.6153 | 0.0127 | |
| SRP064894 | CLTA | 1211 | RNAseq | 0.0754 | 0.7759 | |
| SRP133303 | CLTA | 1211 | RNAseq | -0.2079 | 0.0374 | |
| SRP159526 | CLTA | 1211 | RNAseq | -0.5388 | 0.0148 | |
| SRP193095 | CLTA | 1211 | RNAseq | -0.2916 | 0.1704 | |
| SRP219564 | CLTA | 1211 | RNAseq | 0.0576 | 0.8602 | |
| TCGA | CLTA | 1211 | RNAseq | 0.0353 | 0.5395 |
Upregulated datasets: 0; Downregulated datasets: 0.
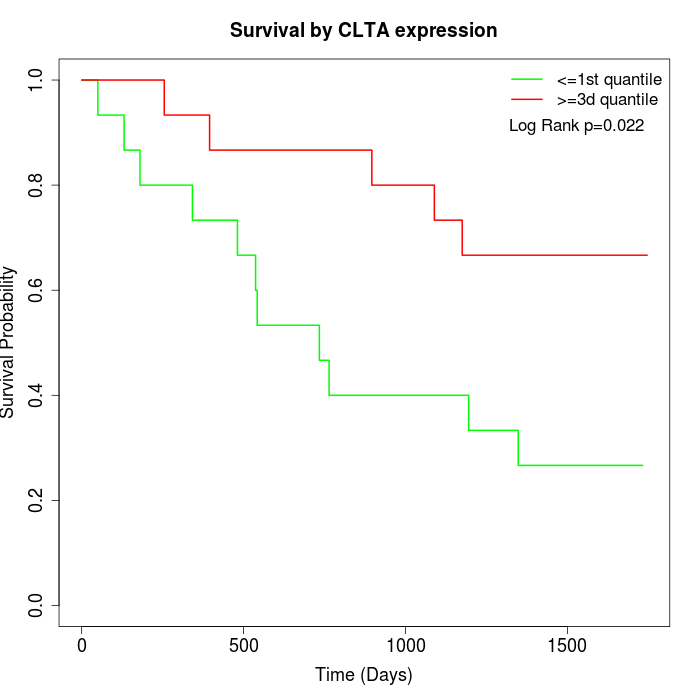
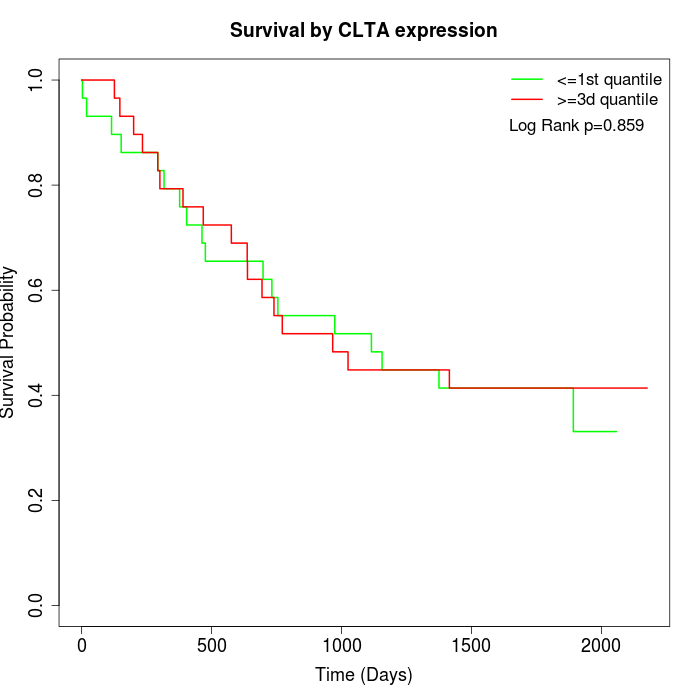
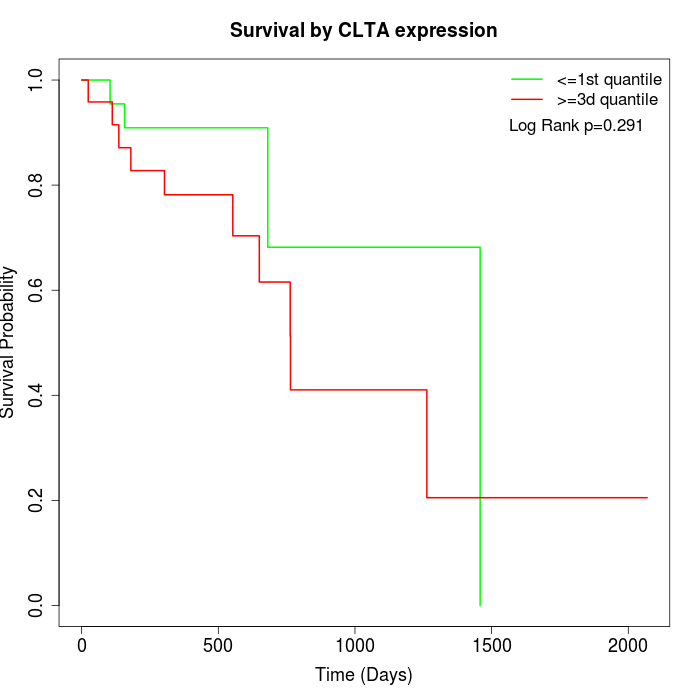
Survival by CLTA expression:
Note: Click image to view full size file.
Copy number change of CLTA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLTA | 1211 | 3 | 11 | 16 | |
| GSE20123 | CLTA | 1211 | 3 | 11 | 16 | |
| GSE43470 | CLTA | 1211 | 3 | 10 | 30 | |
| GSE46452 | CLTA | 1211 | 6 | 15 | 38 | |
| GSE47630 | CLTA | 1211 | 1 | 20 | 19 | |
| GSE54993 | CLTA | 1211 | 6 | 0 | 64 | |
| GSE54994 | CLTA | 1211 | 5 | 12 | 36 | |
| GSE60625 | CLTA | 1211 | 0 | 0 | 11 | |
| GSE74703 | CLTA | 1211 | 2 | 7 | 27 | |
| GSE74704 | CLTA | 1211 | 0 | 9 | 11 | |
| TCGA | CLTA | 1211 | 15 | 44 | 37 |
Total number of gains: 44; Total number of losses: 139; Total Number of normals: 305.
Somatic mutations of CLTA:
Generating mutation plots.
Highly correlated genes for CLTA:
Showing top 20/437 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLTA | LDOC1 | 0.771823 | 3 | 0 | 3 |
| CLTA | ABHD14B | 0.751415 | 4 | 0 | 4 |
| CLTA | STK19 | 0.751274 | 3 | 0 | 3 |
| CLTA | C2orf16 | 0.733313 | 3 | 0 | 3 |
| CLTA | PPID | 0.731704 | 3 | 0 | 3 |
| CLTA | KIAA0556 | 0.730067 | 3 | 0 | 3 |
| CLTA | HDHD2 | 0.729794 | 3 | 0 | 3 |
| CLTA | CCDC12 | 0.729508 | 3 | 0 | 3 |
| CLTA | SCFD2 | 0.724681 | 3 | 0 | 3 |
| CLTA | NLGN3 | 0.720624 | 3 | 0 | 3 |
| CLTA | CGNL1 | 0.716441 | 3 | 0 | 3 |
| CLTA | ZNF441 | 0.71578 | 3 | 0 | 3 |
| CLTA | FUCA1 | 0.712379 | 3 | 0 | 3 |
| CLTA | RAB5A | 0.710693 | 3 | 0 | 3 |
| CLTA | PPM1L | 0.708894 | 3 | 0 | 3 |
| CLTA | TMEM218 | 0.708308 | 4 | 0 | 4 |
| CLTA | EIF3D | 0.701354 | 3 | 0 | 3 |
| CLTA | RAB3D | 0.701279 | 3 | 0 | 3 |
| CLTA | OS9 | 0.699081 | 4 | 0 | 3 |
| CLTA | ADAMTS8 | 0.697055 | 3 | 0 | 3 |
For details and further investigation, click here