| Full name: cornichon family AMPA receptor auxiliary protein 3 | Alias Symbol: FLJ38993|CNIH-3 | ||
| Type: protein-coding gene | Cytoband: 1q42.12 | ||
| Entrez ID: 149111 | HGNC ID: HGNC:26802 | Ensembl Gene: ENSG00000143786 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CNIH3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CNIH3 | 149111 | 238503_at | 0.0009 | 0.9973 | |
| GSE20347 | CNIH3 | 149111 | 214841_at | 0.2130 | 0.1968 | |
| GSE23400 | CNIH3 | 149111 | 214841_at | 0.1180 | 0.0376 | |
| GSE26886 | CNIH3 | 149111 | 214841_at | 1.3927 | 0.0112 | |
| GSE29001 | CNIH3 | 149111 | 214841_at | 0.6438 | 0.2007 | |
| GSE38129 | CNIH3 | 149111 | 214841_at | 0.4068 | 0.0150 | |
| GSE45670 | CNIH3 | 149111 | 238503_at | -0.1736 | 0.0473 | |
| GSE53622 | CNIH3 | 149111 | 54743 | 1.6352 | 0.0000 | |
| GSE53624 | CNIH3 | 149111 | 54743 | 1.8504 | 0.0000 | |
| GSE63941 | CNIH3 | 149111 | 214841_at | -0.9871 | 0.1343 | |
| GSE77861 | CNIH3 | 149111 | 238503_at | -0.1395 | 0.1645 | |
| GSE97050 | CNIH3 | 149111 | A_23_P384044 | 0.0670 | 0.8717 | |
| SRP064894 | CNIH3 | 149111 | RNAseq | 0.6473 | 0.0443 | |
| SRP133303 | CNIH3 | 149111 | RNAseq | 0.4295 | 0.2295 | |
| SRP159526 | CNIH3 | 149111 | RNAseq | 0.3582 | 0.5077 | |
| SRP193095 | CNIH3 | 149111 | RNAseq | 0.2946 | 0.1853 | |
| SRP219564 | CNIH3 | 149111 | RNAseq | -0.2345 | 0.6519 | |
| TCGA | CNIH3 | 149111 | RNAseq | 0.1827 | 0.5326 |
Upregulated datasets: 3; Downregulated datasets: 0.
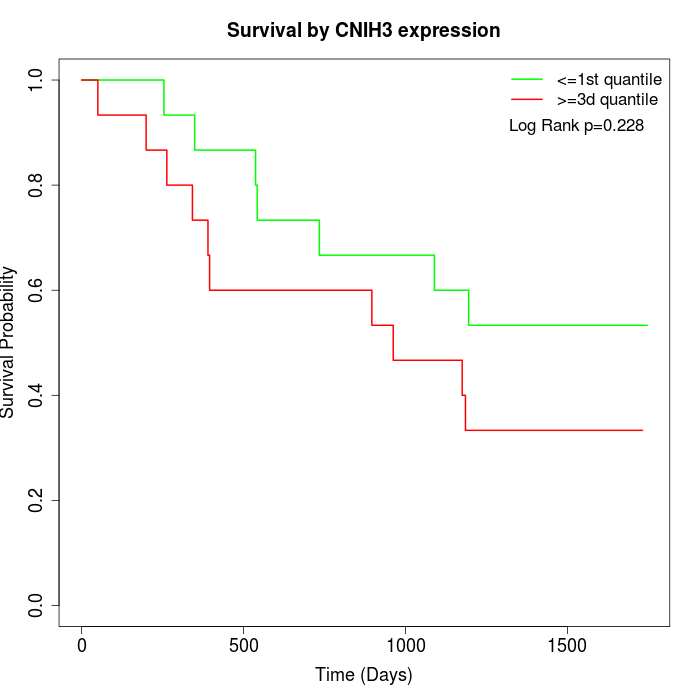
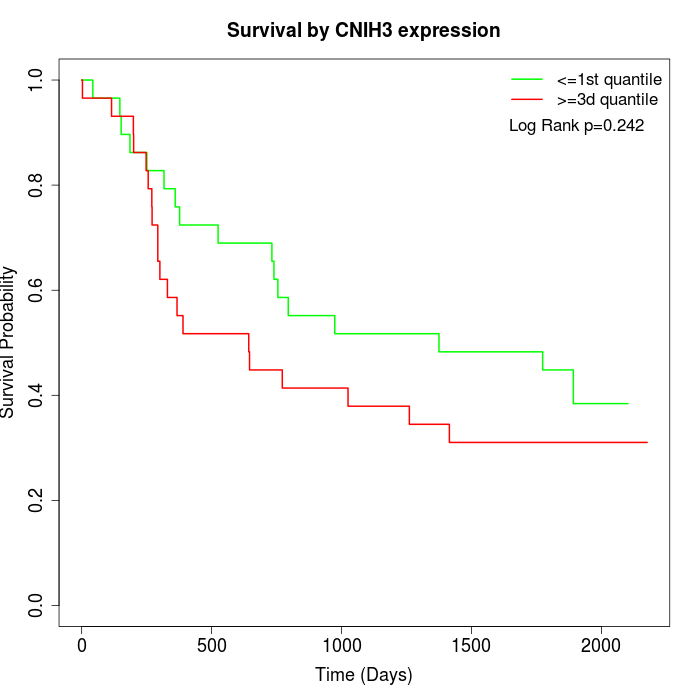
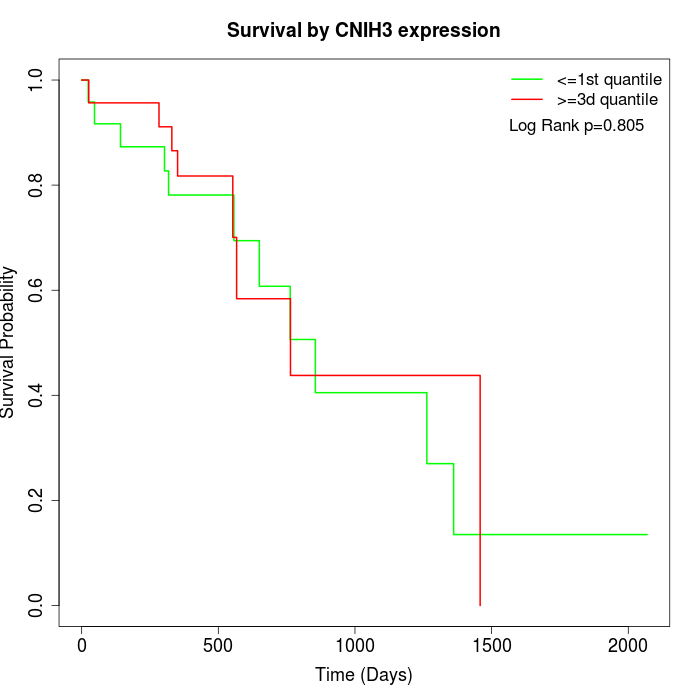
Survival by CNIH3 expression:
Note: Click image to view full size file.
Copy number change of CNIH3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CNIH3 | 149111 | 10 | 0 | 20 | |
| GSE20123 | CNIH3 | 149111 | 10 | 0 | 20 | |
| GSE43470 | CNIH3 | 149111 | 7 | 1 | 35 | |
| GSE46452 | CNIH3 | 149111 | 3 | 1 | 55 | |
| GSE47630 | CNIH3 | 149111 | 15 | 0 | 25 | |
| GSE54993 | CNIH3 | 149111 | 0 | 6 | 64 | |
| GSE54994 | CNIH3 | 149111 | 16 | 0 | 37 | |
| GSE60625 | CNIH3 | 149111 | 0 | 0 | 11 | |
| GSE74703 | CNIH3 | 149111 | 7 | 1 | 28 | |
| GSE74704 | CNIH3 | 149111 | 4 | 0 | 16 | |
| TCGA | CNIH3 | 149111 | 45 | 3 | 48 |
Total number of gains: 117; Total number of losses: 12; Total Number of normals: 359.
Somatic mutations of CNIH3:
Generating mutation plots.
Highly correlated genes for CNIH3:
Showing top 20/188 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CNIH3 | FGF3 | 0.799484 | 3 | 0 | 3 |
| CNIH3 | ABCA3 | 0.789977 | 3 | 0 | 3 |
| CNIH3 | MEX3B | 0.763851 | 4 | 0 | 4 |
| CNIH3 | FKBP7 | 0.759155 | 4 | 0 | 4 |
| CNIH3 | RAMP1 | 0.751272 | 3 | 0 | 3 |
| CNIH3 | CSRNP2 | 0.750508 | 3 | 0 | 3 |
| CNIH3 | COL8A2 | 0.741939 | 3 | 0 | 3 |
| CNIH3 | CEP250 | 0.738485 | 3 | 0 | 3 |
| CNIH3 | OSGEP | 0.72375 | 3 | 0 | 3 |
| CNIH3 | PSEN2 | 0.723128 | 3 | 0 | 3 |
| CNIH3 | SPIC | 0.722642 | 3 | 0 | 3 |
| CNIH3 | ZNF124 | 0.720531 | 3 | 0 | 3 |
| CNIH3 | CTPS2 | 0.715784 | 3 | 0 | 3 |
| CNIH3 | SPON2 | 0.71569 | 3 | 0 | 3 |
| CNIH3 | GXYLT2 | 0.710629 | 3 | 0 | 3 |
| CNIH3 | FAM117A | 0.705641 | 3 | 0 | 3 |
| CNIH3 | PIK3R3 | 0.705376 | 3 | 0 | 3 |
| CNIH3 | RAB34 | 0.701574 | 3 | 0 | 3 |
| CNIH3 | FADS1 | 0.695918 | 3 | 0 | 3 |
| CNIH3 | MTMR4 | 0.695123 | 3 | 0 | 3 |
For details and further investigation, click here