| Full name: cornichon family AMPA receptor auxiliary protein 4 | Alias Symbol: HSPC163 | ||
| Type: protein-coding gene | Cytoband: 1q42.11 | ||
| Entrez ID: 29097 | HGNC ID: HGNC:25013 | Ensembl Gene: ENSG00000143771 | OMIM ID: 617483 |
| Drug and gene relationship at DGIdb | |||
Expression of CNIH4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CNIH4 | 29097 | 223993_s_at | 0.5238 | 0.1059 | |
| GSE20347 | CNIH4 | 29097 | 218728_s_at | 0.4594 | 0.0014 | |
| GSE23400 | CNIH4 | 29097 | 218728_s_at | 0.8144 | 0.0000 | |
| GSE26886 | CNIH4 | 29097 | 223993_s_at | -0.0455 | 0.8738 | |
| GSE29001 | CNIH4 | 29097 | 218728_s_at | 0.5591 | 0.0593 | |
| GSE38129 | CNIH4 | 29097 | 218728_s_at | 0.7658 | 0.0000 | |
| GSE45670 | CNIH4 | 29097 | 223993_s_at | 0.4877 | 0.0002 | |
| GSE53622 | CNIH4 | 29097 | 27586 | 0.4829 | 0.0000 | |
| GSE53624 | CNIH4 | 29097 | 27586 | 0.8051 | 0.0000 | |
| GSE63941 | CNIH4 | 29097 | 223993_s_at | 0.7924 | 0.0067 | |
| GSE77861 | CNIH4 | 29097 | 223993_s_at | 0.4100 | 0.0202 | |
| GSE97050 | CNIH4 | 29097 | A_23_P200507 | 0.1779 | 0.5484 | |
| SRP007169 | CNIH4 | 29097 | RNAseq | -0.2729 | 0.4693 | |
| SRP008496 | CNIH4 | 29097 | RNAseq | -0.2479 | 0.4526 | |
| SRP064894 | CNIH4 | 29097 | RNAseq | 0.4398 | 0.0375 | |
| SRP133303 | CNIH4 | 29097 | RNAseq | 0.7244 | 0.0001 | |
| SRP159526 | CNIH4 | 29097 | RNAseq | 0.3022 | 0.4800 | |
| SRP193095 | CNIH4 | 29097 | RNAseq | 0.0170 | 0.9132 | |
| SRP219564 | CNIH4 | 29097 | RNAseq | 0.2252 | 0.5539 | |
| TCGA | CNIH4 | 29097 | RNAseq | 0.2174 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
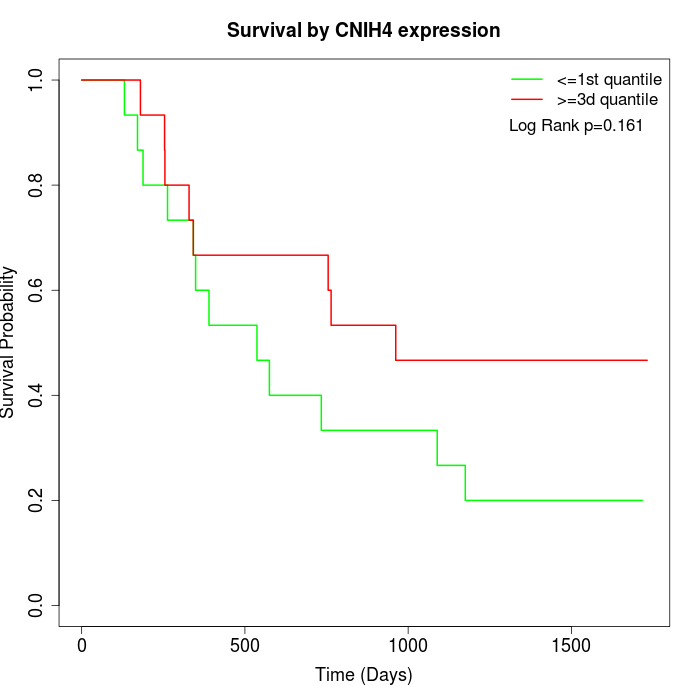
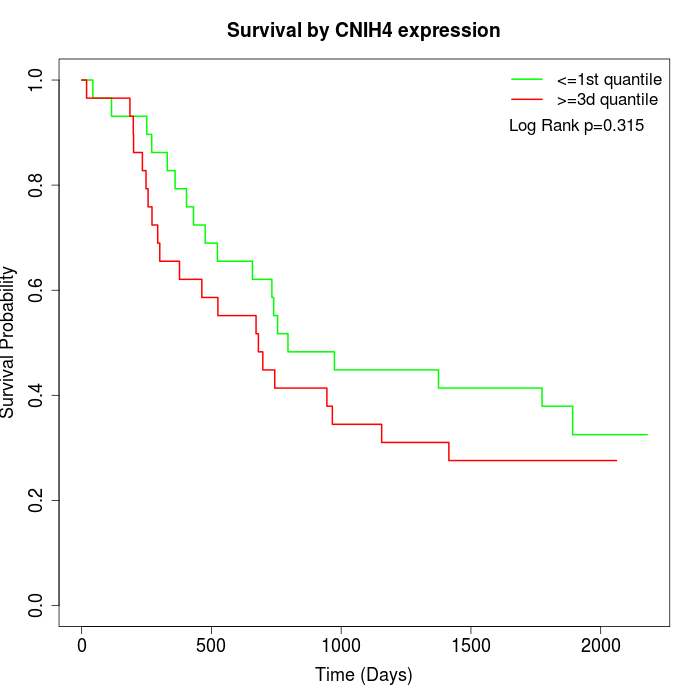
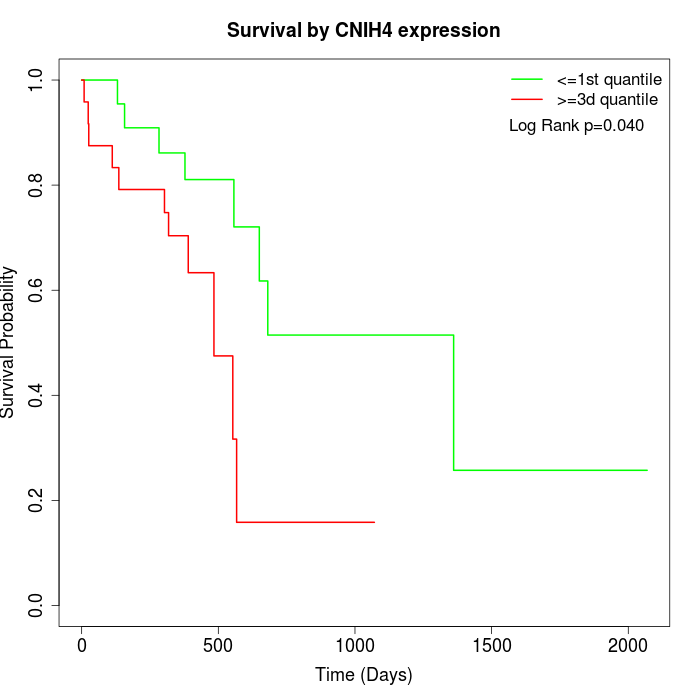
Survival by CNIH4 expression:
Note: Click image to view full size file.
Copy number change of CNIH4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CNIH4 | 29097 | 10 | 0 | 20 | |
| GSE20123 | CNIH4 | 29097 | 10 | 0 | 20 | |
| GSE43470 | CNIH4 | 29097 | 7 | 0 | 36 | |
| GSE46452 | CNIH4 | 29097 | 3 | 1 | 55 | |
| GSE47630 | CNIH4 | 29097 | 15 | 0 | 25 | |
| GSE54993 | CNIH4 | 29097 | 0 | 6 | 64 | |
| GSE54994 | CNIH4 | 29097 | 16 | 0 | 37 | |
| GSE60625 | CNIH4 | 29097 | 0 | 0 | 11 | |
| GSE74703 | CNIH4 | 29097 | 7 | 0 | 29 | |
| GSE74704 | CNIH4 | 29097 | 4 | 0 | 16 | |
| TCGA | CNIH4 | 29097 | 46 | 3 | 47 |
Total number of gains: 118; Total number of losses: 10; Total Number of normals: 360.
Somatic mutations of CNIH4:
Generating mutation plots.
Highly correlated genes for CNIH4:
Showing top 20/1689 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CNIH4 | EBNA1BP2 | 0.786298 | 3 | 0 | 3 |
| CNIH4 | PYCR2 | 0.783769 | 3 | 0 | 3 |
| CNIH4 | CCDC85C | 0.778856 | 3 | 0 | 3 |
| CNIH4 | FADS1 | 0.74601 | 3 | 0 | 3 |
| CNIH4 | RAB34 | 0.734959 | 3 | 0 | 3 |
| CNIH4 | C1orf131 | 0.730961 | 8 | 0 | 8 |
| CNIH4 | TMEM138 | 0.725727 | 7 | 0 | 7 |
| CNIH4 | CPSF6 | 0.722955 | 11 | 0 | 11 |
| CNIH4 | SCNM1 | 0.721588 | 3 | 0 | 3 |
| CNIH4 | TBRG4 | 0.719257 | 3 | 0 | 3 |
| CNIH4 | PSMD14 | 0.719054 | 11 | 0 | 10 |
| CNIH4 | NTMT1 | 0.718881 | 5 | 0 | 5 |
| CNIH4 | EFTUD2 | 0.71799 | 7 | 0 | 7 |
| CNIH4 | WDR54 | 0.717956 | 6 | 0 | 6 |
| CNIH4 | TAF2 | 0.715905 | 9 | 0 | 9 |
| CNIH4 | AURKB | 0.714668 | 9 | 0 | 9 |
| CNIH4 | KRTCAP2 | 0.71178 | 4 | 0 | 3 |
| CNIH4 | FSCN1 | 0.71039 | 9 | 0 | 8 |
| CNIH4 | HEATR1 | 0.706951 | 12 | 0 | 11 |
| CNIH4 | CKS1B | 0.70589 | 11 | 0 | 10 |
For details and further investigation, click here