| Full name: cAMP responsive element modulator | Alias Symbol: hCREM-2 | ||
| Type: protein-coding gene | Cytoband: 10p11.21 | ||
| Entrez ID: 1390 | HGNC ID: HGNC:2352 | Ensembl Gene: ENSG00000095794 | OMIM ID: 123812 |
| Drug and gene relationship at DGIdb | |||
CREM involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa05166 | HTLV-I infection |
Expression of CREM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CREM | 1390 | 207630_s_at | 0.2947 | 0.5315 | |
| GSE20347 | CREM | 1390 | 209967_s_at | 0.2892 | 0.1661 | |
| GSE23400 | CREM | 1390 | 214508_x_at | -0.0132 | 0.8347 | |
| GSE26886 | CREM | 1390 | 209967_s_at | 0.9144 | 0.0019 | |
| GSE29001 | CREM | 1390 | 209967_s_at | 0.9718 | 0.0162 | |
| GSE38129 | CREM | 1390 | 209967_s_at | 0.0055 | 0.9859 | |
| GSE45670 | CREM | 1390 | 209967_s_at | 0.0219 | 0.9553 | |
| GSE53622 | CREM | 1390 | 30878 | -0.0401 | 0.7833 | |
| GSE53624 | CREM | 1390 | 30878 | 0.3186 | 0.0008 | |
| GSE63941 | CREM | 1390 | 207630_s_at | -0.3310 | 0.7207 | |
| GSE77861 | CREM | 1390 | 214508_x_at | 0.1864 | 0.3408 | |
| GSE97050 | CREM | 1390 | A_23_P201979 | 0.1016 | 0.7371 | |
| SRP007169 | CREM | 1390 | RNAseq | 0.6933 | 0.0819 | |
| SRP008496 | CREM | 1390 | RNAseq | 1.1492 | 0.0011 | |
| SRP064894 | CREM | 1390 | RNAseq | 0.3083 | 0.1759 | |
| SRP133303 | CREM | 1390 | RNAseq | 0.9017 | 0.0014 | |
| SRP159526 | CREM | 1390 | RNAseq | 0.2580 | 0.4553 | |
| SRP193095 | CREM | 1390 | RNAseq | 0.4969 | 0.0005 | |
| SRP219564 | CREM | 1390 | RNAseq | 0.3636 | 0.3929 | |
| TCGA | CREM | 1390 | RNAseq | -0.1117 | 0.1388 |
Upregulated datasets: 1; Downregulated datasets: 0.
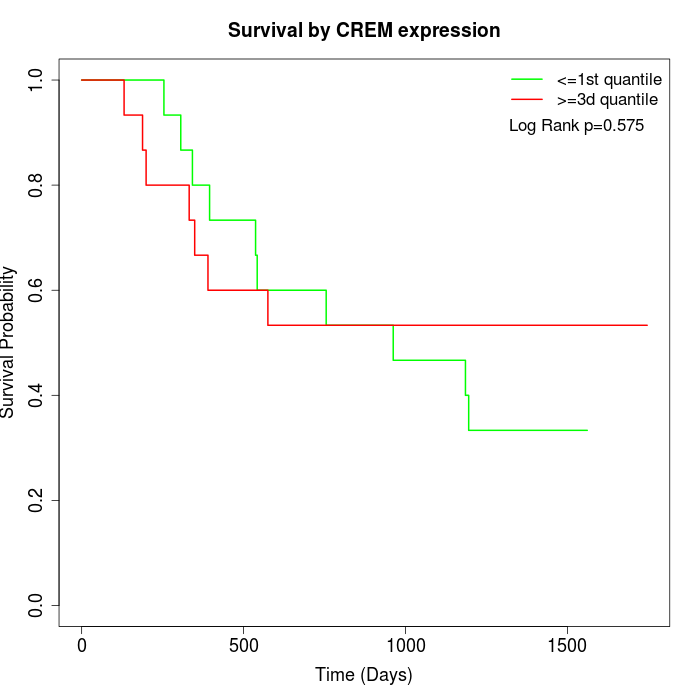
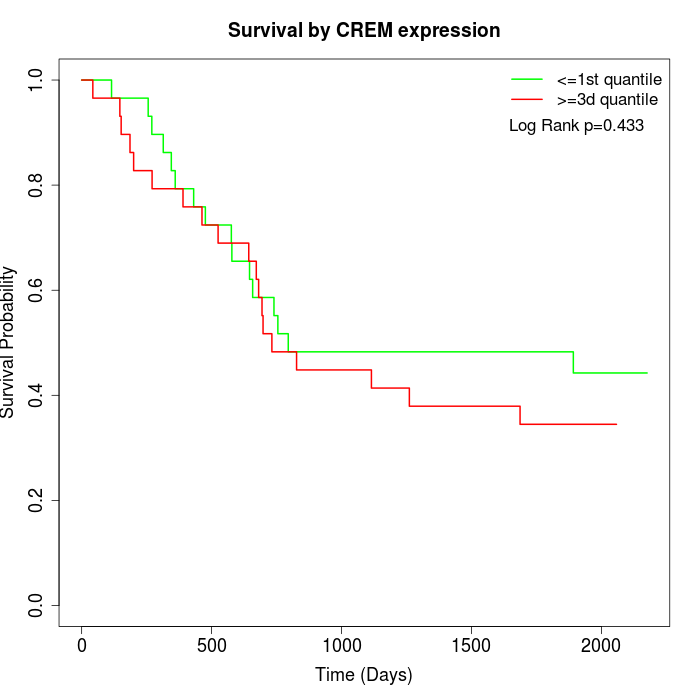
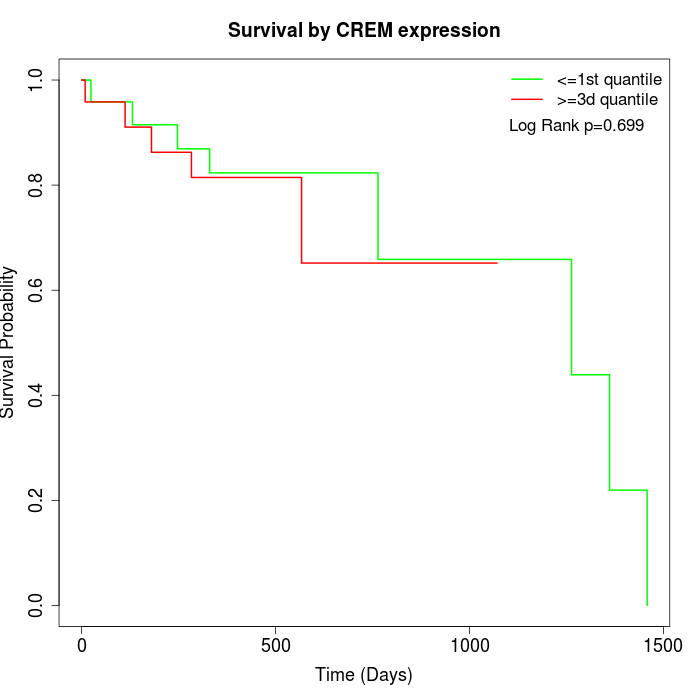
Survival by CREM expression:
Note: Click image to view full size file.
Copy number change of CREM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CREM | 1390 | 4 | 7 | 19 | |
| GSE20123 | CREM | 1390 | 4 | 6 | 20 | |
| GSE43470 | CREM | 1390 | 3 | 3 | 37 | |
| GSE46452 | CREM | 1390 | 2 | 13 | 44 | |
| GSE47630 | CREM | 1390 | 7 | 10 | 23 | |
| GSE54993 | CREM | 1390 | 8 | 1 | 61 | |
| GSE54994 | CREM | 1390 | 2 | 10 | 41 | |
| GSE60625 | CREM | 1390 | 0 | 0 | 11 | |
| GSE74703 | CREM | 1390 | 2 | 1 | 33 | |
| GSE74704 | CREM | 1390 | 0 | 5 | 15 | |
| TCGA | CREM | 1390 | 17 | 24 | 55 |
Total number of gains: 49; Total number of losses: 80; Total Number of normals: 359.
Somatic mutations of CREM:
Generating mutation plots.
Highly correlated genes for CREM:
Showing top 20/903 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CREM | POLR2B | 0.735799 | 3 | 0 | 3 |
| CREM | C3orf33 | 0.733999 | 3 | 0 | 3 |
| CREM | PRELID2 | 0.722818 | 3 | 0 | 3 |
| CREM | HIC1 | 0.717825 | 5 | 0 | 5 |
| CREM | UBA2 | 0.711793 | 3 | 0 | 3 |
| CREM | ZFHX3 | 0.701913 | 3 | 0 | 3 |
| CREM | FXR1 | 0.700011 | 4 | 0 | 4 |
| CREM | C19orf38 | 0.695455 | 3 | 0 | 3 |
| CREM | ACTL6A | 0.691462 | 4 | 0 | 4 |
| CREM | CNIH3 | 0.691355 | 3 | 0 | 3 |
| CREM | SULT1A2 | 0.690379 | 3 | 0 | 3 |
| CREM | MRPL47 | 0.690328 | 3 | 0 | 3 |
| CREM | ZNF827 | 0.689133 | 3 | 0 | 3 |
| CREM | CCDC59 | 0.688013 | 4 | 0 | 3 |
| CREM | GRAMD1A | 0.686063 | 5 | 0 | 5 |
| CREM | ZDHHC7 | 0.684169 | 3 | 0 | 3 |
| CREM | IL33 | 0.682896 | 3 | 0 | 3 |
| CREM | RAB8B | 0.681059 | 5 | 0 | 5 |
| CREM | TEAD2 | 0.680657 | 4 | 0 | 3 |
| CREM | KCNAB2 | 0.679474 | 4 | 0 | 3 |
For details and further investigation, click here