| Full name: potassium voltage-gated channel subfamily A regulatory beta subunit 2 | Alias Symbol: AKR6A5|KCNA2B|HKvbeta2.1|HKvbeta2.2 | ||
| Type: protein-coding gene | Cytoband: 1p36.31 | ||
| Entrez ID: 8514 | HGNC ID: HGNC:6229 | Ensembl Gene: ENSG00000069424 | OMIM ID: 601142 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNAB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNAB2 | 8514 | 203402_at | 0.2877 | 0.6014 | |
| GSE20347 | KCNAB2 | 8514 | 203402_at | 0.2782 | 0.0187 | |
| GSE23400 | KCNAB2 | 8514 | 203402_at | 0.0674 | 0.0933 | |
| GSE26886 | KCNAB2 | 8514 | 203402_at | 0.5820 | 0.0001 | |
| GSE29001 | KCNAB2 | 8514 | 203402_at | 0.0703 | 0.7826 | |
| GSE38129 | KCNAB2 | 8514 | 203402_at | 0.2121 | 0.0860 | |
| GSE45670 | KCNAB2 | 8514 | 203402_at | -0.1021 | 0.4245 | |
| GSE53622 | KCNAB2 | 8514 | 31363 | 0.6736 | 0.0000 | |
| GSE53624 | KCNAB2 | 8514 | 31363 | 0.9041 | 0.0000 | |
| GSE63941 | KCNAB2 | 8514 | 203402_at | 0.3247 | 0.4229 | |
| GSE77861 | KCNAB2 | 8514 | 203402_at | 0.1024 | 0.6569 | |
| GSE97050 | KCNAB2 | 8514 | A_24_P151 | 0.7358 | 0.0892 | |
| SRP007169 | KCNAB2 | 8514 | RNAseq | 1.7490 | 0.0123 | |
| SRP008496 | KCNAB2 | 8514 | RNAseq | 1.1895 | 0.0070 | |
| SRP064894 | KCNAB2 | 8514 | RNAseq | 0.9844 | 0.0006 | |
| SRP133303 | KCNAB2 | 8514 | RNAseq | 0.3583 | 0.0986 | |
| SRP159526 | KCNAB2 | 8514 | RNAseq | 0.1506 | 0.7464 | |
| SRP193095 | KCNAB2 | 8514 | RNAseq | 1.0106 | 0.0000 | |
| SRP219564 | KCNAB2 | 8514 | RNAseq | 0.3599 | 0.4194 | |
| TCGA | KCNAB2 | 8514 | RNAseq | 0.0636 | 0.5940 |
Upregulated datasets: 3; Downregulated datasets: 0.
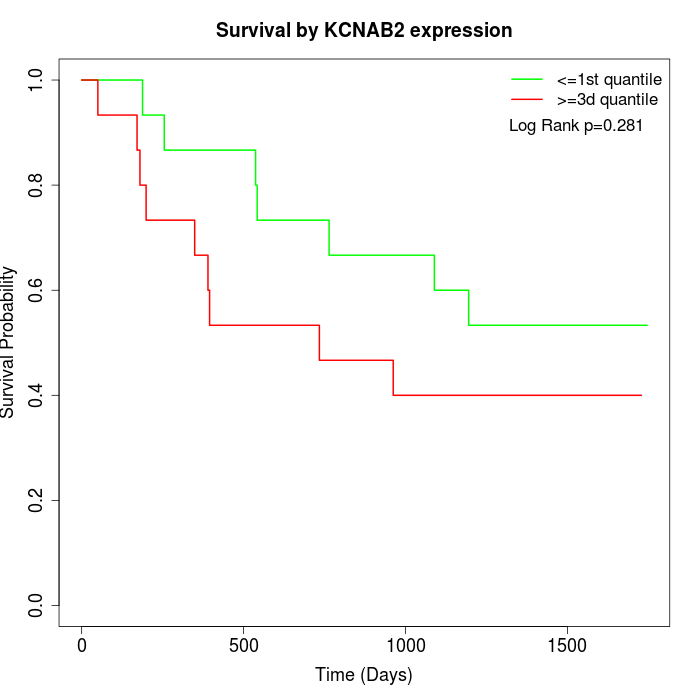
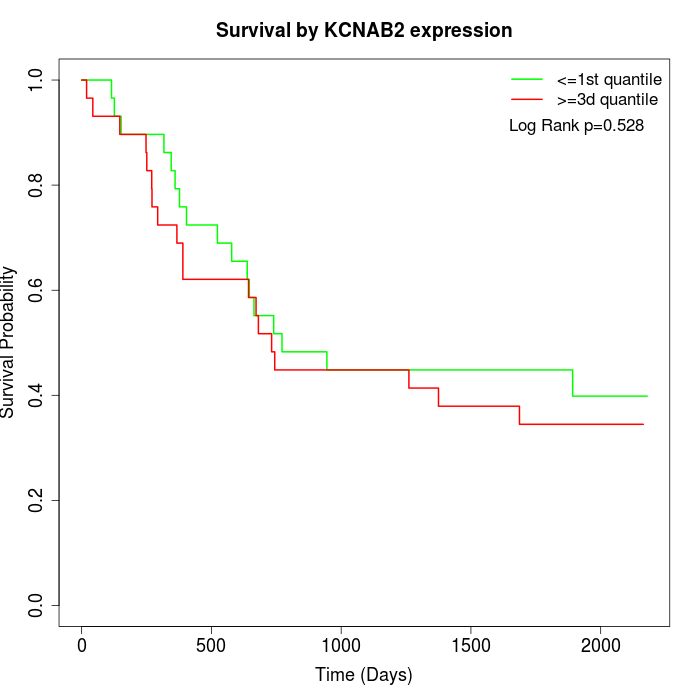
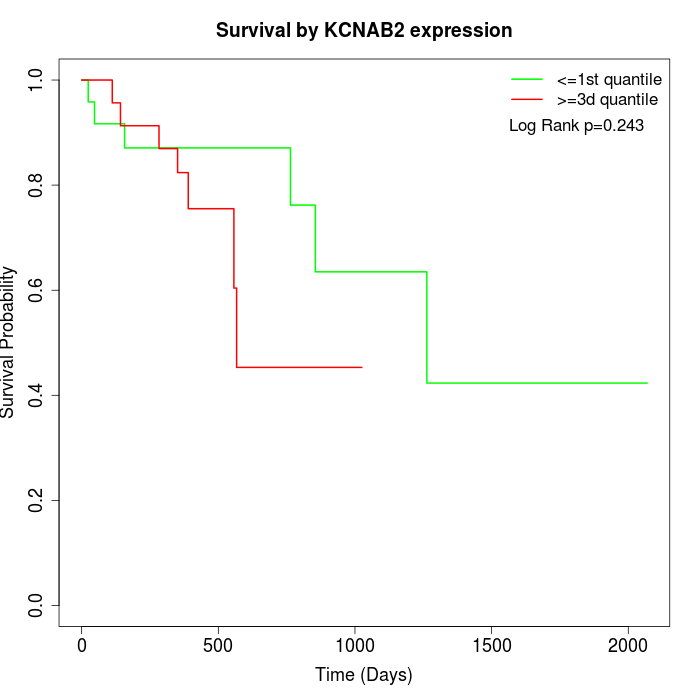
Survival by KCNAB2 expression:
Note: Click image to view full size file.
Copy number change of KCNAB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNAB2 | 8514 | 3 | 6 | 21 | |
| GSE20123 | KCNAB2 | 8514 | 3 | 5 | 22 | |
| GSE43470 | KCNAB2 | 8514 | 6 | 5 | 32 | |
| GSE46452 | KCNAB2 | 8514 | 7 | 1 | 51 | |
| GSE47630 | KCNAB2 | 8514 | 8 | 4 | 28 | |
| GSE54993 | KCNAB2 | 8514 | 3 | 2 | 65 | |
| GSE54994 | KCNAB2 | 8514 | 14 | 3 | 36 | |
| GSE60625 | KCNAB2 | 8514 | 0 | 0 | 11 | |
| GSE74703 | KCNAB2 | 8514 | 5 | 3 | 28 | |
| GSE74704 | KCNAB2 | 8514 | 3 | 0 | 17 | |
| TCGA | KCNAB2 | 8514 | 12 | 22 | 62 |
Total number of gains: 64; Total number of losses: 51; Total Number of normals: 373.
Somatic mutations of KCNAB2:
Generating mutation plots.
Highly correlated genes for KCNAB2:
Showing top 20/1090 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNAB2 | KHSRP | 0.815053 | 3 | 0 | 3 |
| KCNAB2 | TRIM47 | 0.811543 | 3 | 0 | 3 |
| KCNAB2 | GNGT2 | 0.801937 | 3 | 0 | 3 |
| KCNAB2 | SLC9A1 | 0.783388 | 3 | 0 | 3 |
| KCNAB2 | FCGR3A | 0.772972 | 3 | 0 | 3 |
| KCNAB2 | A1BG | 0.77034 | 3 | 0 | 3 |
| KCNAB2 | ITPK1 | 0.766092 | 3 | 0 | 3 |
| KCNAB2 | CERKL | 0.762393 | 3 | 0 | 3 |
| KCNAB2 | CACNA1B | 0.75511 | 3 | 0 | 3 |
| KCNAB2 | SUGP1 | 0.752539 | 3 | 0 | 3 |
| KCNAB2 | YIPF2 | 0.744698 | 3 | 0 | 3 |
| KCNAB2 | ITPR3 | 0.741371 | 3 | 0 | 3 |
| KCNAB2 | CTBP2 | 0.736758 | 3 | 0 | 3 |
| KCNAB2 | MZF1 | 0.731703 | 3 | 0 | 3 |
| KCNAB2 | KCTD13 | 0.731494 | 5 | 0 | 5 |
| KCNAB2 | SSBP4 | 0.731437 | 4 | 0 | 4 |
| KCNAB2 | CCDC86 | 0.728025 | 3 | 0 | 3 |
| KCNAB2 | LRRC8C | 0.727481 | 3 | 0 | 3 |
| KCNAB2 | HGF | 0.727217 | 4 | 0 | 4 |
| KCNAB2 | PPP1R35 | 0.727027 | 3 | 0 | 3 |
For details and further investigation, click here