| Full name: carnitine O-octanoyltransferase | Alias Symbol: COT | ||
| Type: protein-coding gene | Cytoband: 7q21.12 | ||
| Entrez ID: 54677 | HGNC ID: HGNC:2366 | Ensembl Gene: ENSG00000005469 | OMIM ID: 606090 |
| Drug and gene relationship at DGIdb | |||
Expression of CROT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CROT | 54677 | 204573_at | -0.7224 | 0.3469 | |
| GSE20347 | CROT | 54677 | 204573_at | -1.0450 | 0.0000 | |
| GSE23400 | CROT | 54677 | 204573_at | -0.3767 | 0.0000 | |
| GSE26886 | CROT | 54677 | 204573_at | -1.6501 | 0.0000 | |
| GSE29001 | CROT | 54677 | 204573_at | -0.9943 | 0.0001 | |
| GSE38129 | CROT | 54677 | 204573_at | -0.7578 | 0.0015 | |
| GSE45670 | CROT | 54677 | 204573_at | -0.2444 | 0.3425 | |
| GSE53622 | CROT | 54677 | 55429 | -1.0832 | 0.0000 | |
| GSE53624 | CROT | 54677 | 55429 | -1.6698 | 0.0000 | |
| GSE63941 | CROT | 54677 | 204573_at | -2.1326 | 0.0600 | |
| GSE77861 | CROT | 54677 | 204573_at | -0.6323 | 0.0052 | |
| GSE97050 | CROT | 54677 | A_23_P168669 | -0.3586 | 0.2408 | |
| SRP007169 | CROT | 54677 | RNAseq | -2.4701 | 0.0000 | |
| SRP008496 | CROT | 54677 | RNAseq | -1.6438 | 0.0000 | |
| SRP064894 | CROT | 54677 | RNAseq | -1.3499 | 0.0000 | |
| SRP133303 | CROT | 54677 | RNAseq | -0.4363 | 0.0615 | |
| SRP159526 | CROT | 54677 | RNAseq | -1.6748 | 0.0000 | |
| SRP193095 | CROT | 54677 | RNAseq | -0.8850 | 0.0000 | |
| SRP219564 | CROT | 54677 | RNAseq | -0.1561 | 0.7891 | |
| TCGA | CROT | 54677 | RNAseq | 0.0327 | 0.7343 |
Upregulated datasets: 0; Downregulated datasets: 8.
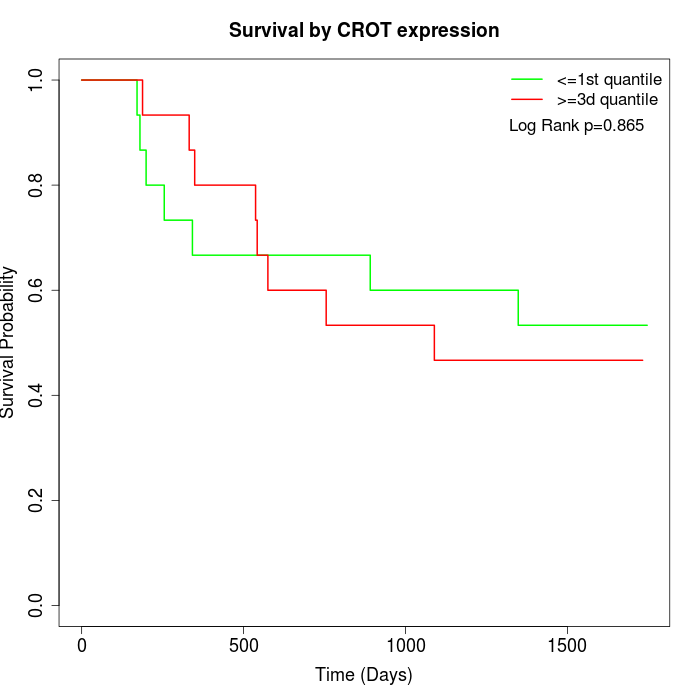
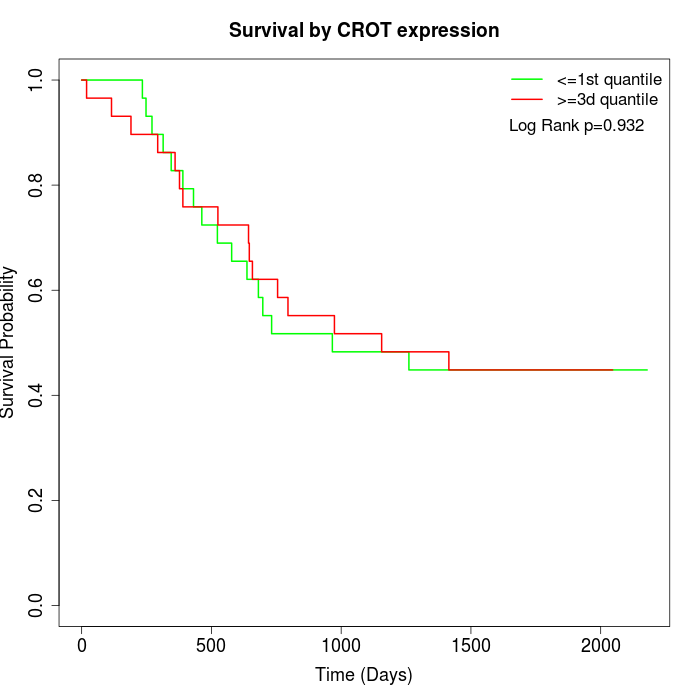
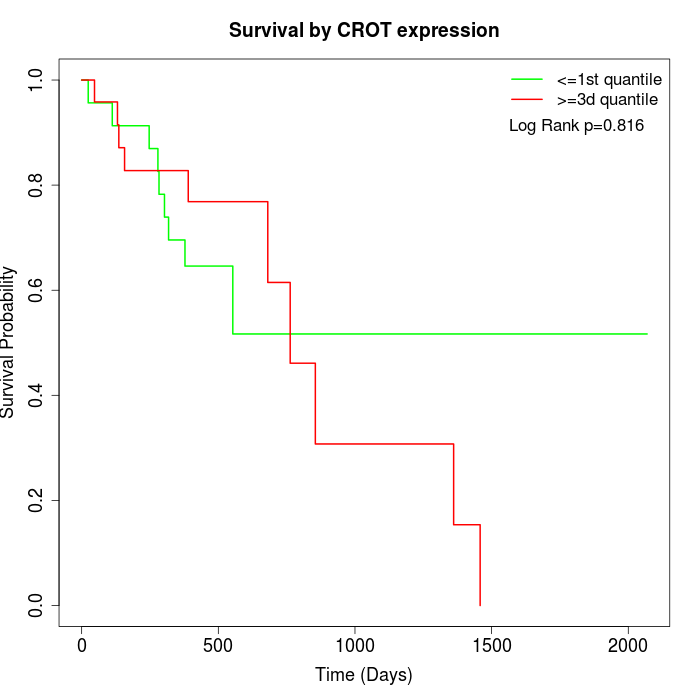
Survival by CROT expression:
Note: Click image to view full size file.
Copy number change of CROT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CROT | 54677 | 13 | 1 | 16 | |
| GSE20123 | CROT | 54677 | 13 | 1 | 16 | |
| GSE43470 | CROT | 54677 | 5 | 1 | 37 | |
| GSE46452 | CROT | 54677 | 12 | 0 | 47 | |
| GSE47630 | CROT | 54677 | 8 | 2 | 30 | |
| GSE54993 | CROT | 54677 | 2 | 7 | 61 | |
| GSE54994 | CROT | 54677 | 17 | 2 | 34 | |
| GSE60625 | CROT | 54677 | 0 | 0 | 11 | |
| GSE74703 | CROT | 54677 | 4 | 1 | 31 | |
| GSE74704 | CROT | 54677 | 9 | 1 | 10 | |
| TCGA | CROT | 54677 | 54 | 4 | 38 |
Total number of gains: 137; Total number of losses: 20; Total Number of normals: 331.
Somatic mutations of CROT:
Generating mutation plots.
Highly correlated genes for CROT:
Showing top 20/815 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CROT | OR2Z1 | 0.807659 | 3 | 0 | 3 |
| CROT | KRTAP20-1 | 0.802827 | 3 | 0 | 3 |
| CROT | FAM163B | 0.80247 | 3 | 0 | 3 |
| CROT | NCCRP1 | 0.79931 | 3 | 0 | 3 |
| CROT | OR10G8 | 0.775217 | 3 | 0 | 3 |
| CROT | CYP3A7 | 0.759869 | 3 | 0 | 3 |
| CROT | MFSD2B | 0.75832 | 3 | 0 | 3 |
| CROT | NANOS2 | 0.757792 | 4 | 0 | 4 |
| CROT | MUC22 | 0.757554 | 3 | 0 | 3 |
| CROT | SIM2 | 0.753767 | 10 | 0 | 10 |
| CROT | SPATA19 | 0.753369 | 4 | 0 | 4 |
| CROT | KRT4 | 0.750535 | 11 | 0 | 10 |
| CROT | MAL | 0.750158 | 11 | 0 | 11 |
| CROT | KRT13 | 0.749086 | 11 | 0 | 11 |
| CROT | CLCA4 | 0.743404 | 11 | 0 | 11 |
| CROT | RAET1E | 0.74196 | 7 | 0 | 7 |
| CROT | NCR1 | 0.741568 | 4 | 0 | 3 |
| CROT | SASH1 | 0.741228 | 11 | 0 | 11 |
| CROT | EPS8L2 | 0.736766 | 12 | 0 | 11 |
| CROT | RIPK4 | 0.735864 | 9 | 0 | 9 |
For details and further investigation, click here