| Full name: chloride channel accessory 4 | Alias Symbol: CaCC2 | ||
| Type: protein-coding gene | Cytoband: 1p22.3 | ||
| Entrez ID: 22802 | HGNC ID: HGNC:2018 | Ensembl Gene: ENSG00000016602 | OMIM ID: 616857 |
| Drug and gene relationship at DGIdb | |||
CLCA4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04972 | Pancreatic secretion |
Expression of CLCA4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLCA4 | 22802 | 220026_at | -4.4537 | 0.0325 | |
| GSE20347 | CLCA4 | 22802 | 220026_at | -5.2707 | 0.0000 | |
| GSE23400 | CLCA4 | 22802 | 220026_at | -4.2447 | 0.0000 | |
| GSE26886 | CLCA4 | 22802 | 220026_at | -7.5902 | 0.0000 | |
| GSE29001 | CLCA4 | 22802 | 220026_at | -5.5407 | 0.0000 | |
| GSE38129 | CLCA4 | 22802 | 220026_at | -3.8592 | 0.0001 | |
| GSE45670 | CLCA4 | 22802 | 220026_at | -1.7721 | 0.0491 | |
| GSE53622 | CLCA4 | 22802 | 16748 | -4.2384 | 0.0000 | |
| GSE53624 | CLCA4 | 22802 | 16748 | -4.8520 | 0.0000 | |
| GSE63941 | CLCA4 | 22802 | 220026_at | 0.2235 | 0.3686 | |
| GSE77861 | CLCA4 | 22802 | 220026_at | -4.0289 | 0.0152 | |
| GSE97050 | CLCA4 | 22802 | A_23_P45751 | -1.7141 | 0.1006 | |
| SRP007169 | CLCA4 | 22802 | RNAseq | -8.7608 | 0.0000 | |
| SRP008496 | CLCA4 | 22802 | RNAseq | -7.7695 | 0.0000 | |
| SRP064894 | CLCA4 | 22802 | RNAseq | -5.2441 | 0.0000 | |
| SRP133303 | CLCA4 | 22802 | RNAseq | -5.2841 | 0.0000 | |
| SRP159526 | CLCA4 | 22802 | RNAseq | -6.6526 | 0.0000 | |
| SRP219564 | CLCA4 | 22802 | RNAseq | -4.7151 | 0.0007 | |
| TCGA | CLCA4 | 22802 | RNAseq | 0.2420 | 0.6431 |
Upregulated datasets: 0; Downregulated datasets: 16.
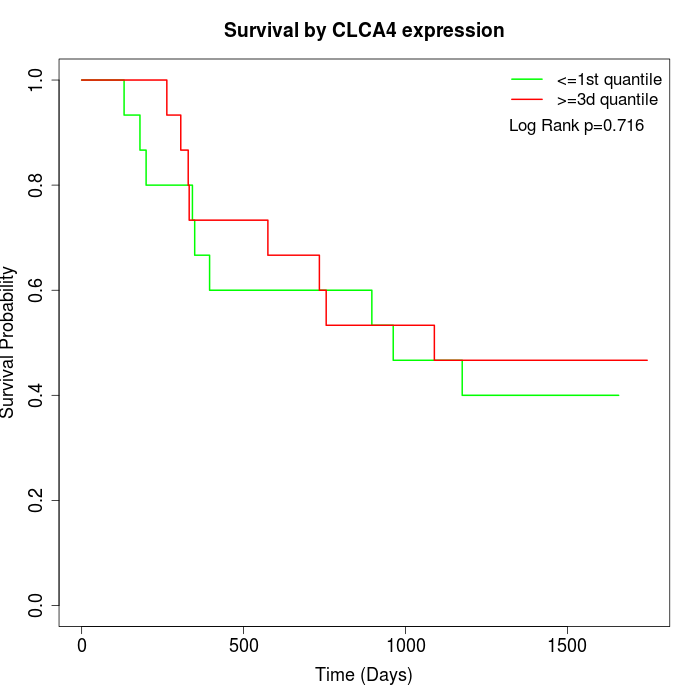
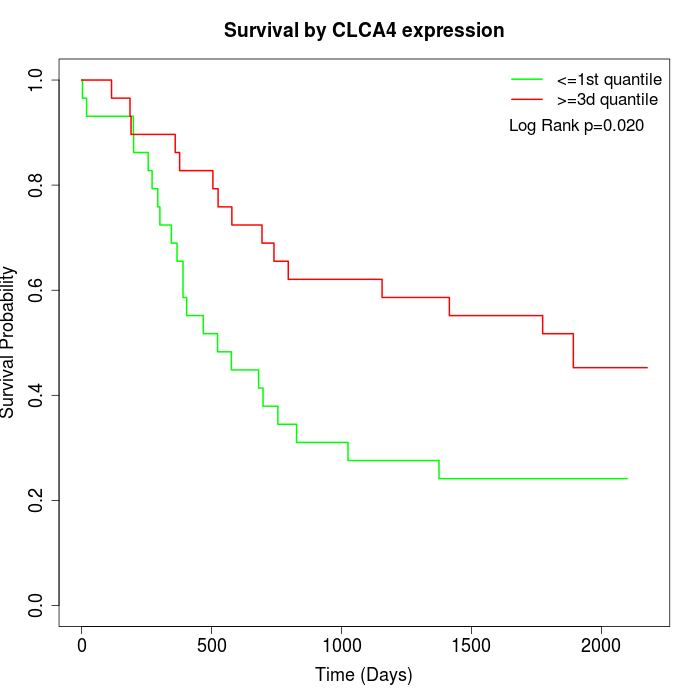
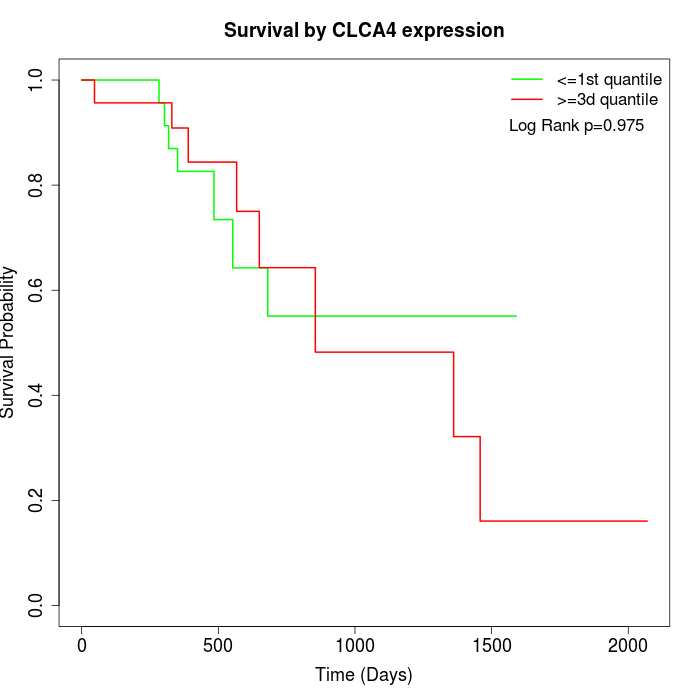
Survival by CLCA4 expression:
Note: Click image to view full size file.
Copy number change of CLCA4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLCA4 | 22802 | 0 | 8 | 22 | |
| GSE20123 | CLCA4 | 22802 | 0 | 8 | 22 | |
| GSE43470 | CLCA4 | 22802 | 2 | 2 | 39 | |
| GSE46452 | CLCA4 | 22802 | 1 | 1 | 57 | |
| GSE47630 | CLCA4 | 22802 | 8 | 5 | 27 | |
| GSE54993 | CLCA4 | 22802 | 0 | 1 | 69 | |
| GSE54994 | CLCA4 | 22802 | 6 | 3 | 44 | |
| GSE60625 | CLCA4 | 22802 | 0 | 0 | 11 | |
| GSE74703 | CLCA4 | 22802 | 1 | 2 | 33 | |
| GSE74704 | CLCA4 | 22802 | 0 | 5 | 15 | |
| TCGA | CLCA4 | 22802 | 6 | 23 | 67 |
Total number of gains: 24; Total number of losses: 58; Total Number of normals: 406.
Somatic mutations of CLCA4:
Generating mutation plots.
Highly correlated genes for CLCA4:
Showing top 20/1549 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLCA4 | TMPRSS11B | 0.932591 | 8 | 0 | 8 |
| CLCA4 | CYSRT1 | 0.924975 | 6 | 0 | 6 |
| CLCA4 | PRSS27 | 0.921577 | 8 | 0 | 8 |
| CLCA4 | CAPN14 | 0.908452 | 7 | 0 | 7 |
| CLCA4 | VSIG10L | 0.897938 | 7 | 0 | 7 |
| CLCA4 | CRNN | 0.890564 | 13 | 0 | 12 |
| CLCA4 | SPINK5 | 0.890223 | 13 | 0 | 12 |
| CLCA4 | TMPRSS11E | 0.886601 | 12 | 0 | 12 |
| CLCA4 | SPINK7 | 0.885371 | 8 | 0 | 8 |
| CLCA4 | TGM3 | 0.883294 | 12 | 0 | 12 |
| CLCA4 | SCEL | 0.879124 | 13 | 0 | 12 |
| CLCA4 | KRT4 | 0.873735 | 13 | 0 | 13 |
| CLCA4 | NCCRP1 | 0.873633 | 4 | 0 | 4 |
| CLCA4 | MAL | 0.870282 | 13 | 0 | 12 |
| CLCA4 | CLIC3 | 0.867301 | 10 | 0 | 10 |
| CLCA4 | ENDOU | 0.863722 | 11 | 0 | 11 |
| CLCA4 | A2ML1 | 0.8607 | 8 | 0 | 8 |
| CLCA4 | NCR1 | 0.860029 | 3 | 0 | 3 |
| CLCA4 | HOPX | 0.859731 | 12 | 0 | 12 |
| CLCA4 | KRT78 | 0.857589 | 8 | 0 | 8 |
For details and further investigation, click here